The human brain is composed of complex circuits of neurons, cells that are specialized to transmit information via electrochemical signals. Like the circuits in a computer, these neuronal circuits must be connected in particular ways to function properly. But with billions of neurons in a single human brain, how does a neuron make the right connections with the right cells?
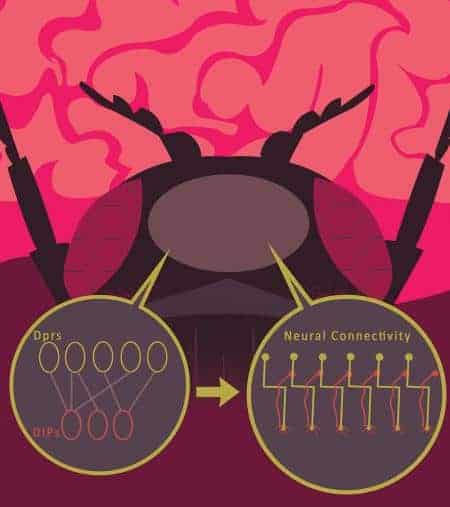
Biologists have long searched for some kind of cellular “identification tags” that label which cells should form connections. Now, researchers from the laboratory of Caltech professor of biology Kai Zinn have identified molecules that act like identification tags on neurons in the fruit fly Drosophila. They discovered that proteins from two different molecular subfamilies, called Dpr and DIP proteins, bind together selectively. This binding can cause neurons that express Dpr proteins to form connections with neurons that express the corresponding DIP protein, playing an important role in directing the development of the neuromuscular and visual systems in growing Drosophila.
A paper detailing the findings is published in the December 17 issue of the journal Cell.
In 2013, a collaboration between Christopher Garcia’s structural biology group at Stanford and the Zinn group at Caltech mapped the interactions between all 200 different Drosophila cell surface proteins. By separating the proteins from the cell and observing their interactions in a test tube, the group determined which proteins bind together. The group developed a complex model of interacting proteins they called the interactome. This work showed that a 21-member subfamily of “immunoglobulin superfamily” proteins, the Dprs, selectively bind to a 9-member subfamily called DIPs.
“Certain members of the Dprs and the DIPs match up and bind together—kind of like a lock and key—in a test tube,” says Zinn. “We wanted to know if they would bind in vivo, in the Drosophila brain, and if that binding would then determine where synapses were formed.”
A synapse is a junction where the wire-like axon of one neuron meets the branched dendrites of another. Information, in the form of chemical signals called neurotransmitters, is passed between neurons across these synapses. “We wanted to know if these interacting proteins on the surface of neuronal cells affected the way that the cells themselves interacted,” says Robert Carrillo, a postdoctoral scholar in the Zinn group and co-first author on the new paper. “We showed that neural cells that expressed matching proteins often formed synapses with each other, and we theorized that the interaction between these molecules was driving the formation of synapses.”
To test this theory, the Zinn group used the well-studied Drosophila visual system to determine the effects of these proteins on development. Neurons in the fly’s eye send axons into layered structures in the visual part of the brain, which is known as the optic lobe. One of these structures, the medulla, is divided into ten layers, and each optic lobe neuron forms synapses within a specific subset of these layers. By removing certain DIP and Dpr proteins in the fly pupa, the researchers caused the axons to “overshoot” their target layers. Additionally, they observed developmental defects in the fly’s neuromuscular system when removing the same proteins. Another paper in the same issue of Cell, from Larry Zipursky’s group at UCLA, also found that expression of Dprs and DIPs correlates with the patterns of synaptic connectivity in the brain.
This finding helps to validate a theory proposed in the 1950s by the late Caltech professor and Nobel Laureate Roger Sperry. Experimenting mostly with fish and frog brains, Sperry discovered that he could manipulate or cut axons between neurons, and the cells would still re-form the right connections.
“Sperry hypothesized that individual neurons must carry some kind of identification tags, whose recognition is used to create the synaptic circuits of the brain,” says Kaushiki Menon, a senior postdoctoral scholar in the Zinn group and a co-first author on the paper. “Our group has shown that the Drosophila Dpr and DIP proteins fit the definition of Sperry’s proposed cellular identification tags.”
Such tinkering with the brain’s circuitry is possible because flies, unlike humans, have brains that are predominantly “hard-wired.” “In mammals, the brain has a basic initial scaffold laid down by genetics, and then over time there is a lot of complicated experience-dependent rearrangement. Essentially, the brain can rewire itself through experience,” Zinn says. “Fly brains can’t do that.”
While their findings are not immediately generalizable to mammals, Zinn and his group hope that they can provide a starting point to probe the structure of the human brain. “We hope that there might be protein networks that function similarly in humans, and these could be relevant to an understanding of how the scaffold of the human brain that exists at birth is assembled through genetics.”
The paper is titled “Control of synaptic connectivity by a network of Drosophila IgSF cell surface proteins.” In addition to Carrillo and Menon, structural biologist Engin Özkan at the University of Chicago is a co-first author. The work was funded by the National Institutes of Health and the Howard Hughes Medical Institute.