wo hundred miles above Earth, NASA has conducted the first genome sequencing in space, and researchers at UC San Francisco helped analyze the data sent back from the International Space Station and confirm that the process was a success.
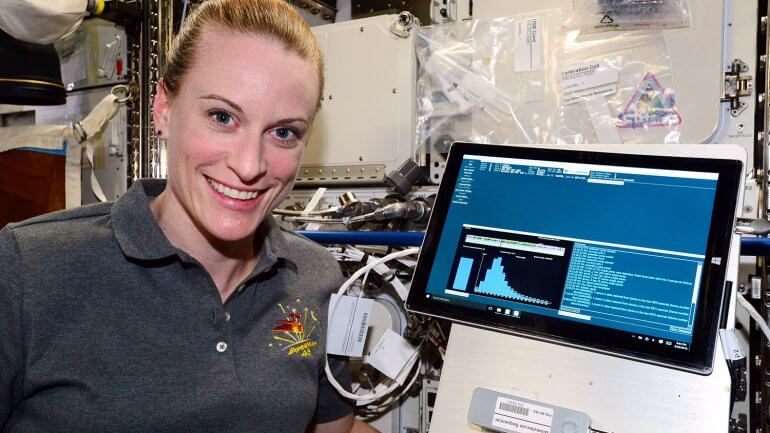
Using a hand-held, USB-powered sequencing device called the MinION, astronaut Kate Rubins, PhD, sequenced samples of mouse, bacteria and virus DNA. This portable sequencing technology could eventually help diagnose sick astronauts, monitor space station food, water, and environment for microbes, and identify extraterrestrial life forms.
As part of a collaboration with NASA on the Biomolecular Sequencer project, Earth-bound researchers – a bicoastal team at UCSF and Weill Cornell Medical College – analyzed the sequencing data from space and compared it to identical samples sequenced on the ground. The analysis would tell whether the journey to space and conditions aboard the space station affected the sequencing results.
Charles Chiu, MD, PhD, director of the UCSF-Abbott Viral Diagnostics and Discovery Center, led the team at UCSF. “Up until recently technology for sequencing in space hasn’t been available because sequencers are generally large bulky instruments,” Chiu said.
The pocket-sized MinION employs a technology known as nanopore sequencing, which sends an ionic current through tiny pores in natural cell membranes. As DNA molecules pass through these nanopores one by one, the changes in ion flow are recorded and used to decode the DNA sequence. A sequencing run can take as little as 10 minutes.
Chiu said a big unknown was whether the microgravity in space would affect the microfluidics of the sequencing device.
“It didn’t turn out to be a huge problem,” Chiu said. His team’s analysis of the space and Earth data found comparable results. “We essentially got equivalent data, and it’s of very high quality, probably within the top 20 percent of nanopore runs that we do routinely here on Earth,” Chiu said.
Besides its portability, another advantage of nanopore sequencing is its ability to rapidly sequence any and all genetic material in a sample, even from organisms that have never been encountered before. Most conventional sequencing methods are confined to a target approach – researchers have to know what they’re looking for and must set up the sequencing accordingly.
Chiu has used nanopore sequencing to test for emerging outbreaks like Ebola and Zika without the need to design a custom diagnostic test. The MinION, made by Oxford Nanopore Technologies, has been employed at field laboratories in some of the remotest areas on Earth – and may someday take a trip to Mars.
In this initial proof-of-principle experiment, the samples were prepared on Earth. On future missions, sample preparation, sequencing, and real-time analysis on a laptop could all be completed in space, and the capacity to do so would be critical for long-term space exploration, Chiu said.
“Onboard sequencing makes it possible for the crew to know what is in their environment at any time,” said project manager and NASA microbiologist Sarah Castro-Wallace, PhD, in a press release. “That allows us on the ground to take appropriate action – do we need to clean this up right away, or will taking antibiotics help or not? We can resupply the station with disinfectants and antibiotics now, but once crews move beyond the station’s low Earth orbit, we need to know when to save those precious resources and when to use them.”