Researchers have developed a new mathematical framework that changes the way we understand the structure of viruses such as Zika and Herpes.
The discovery, by researchers at the University of York (UK) and San Diego State University (US), paves the way for new insights into how viruses form, evolve and infect their hosts and may eventually open up new avenues in anti-viral therapy.
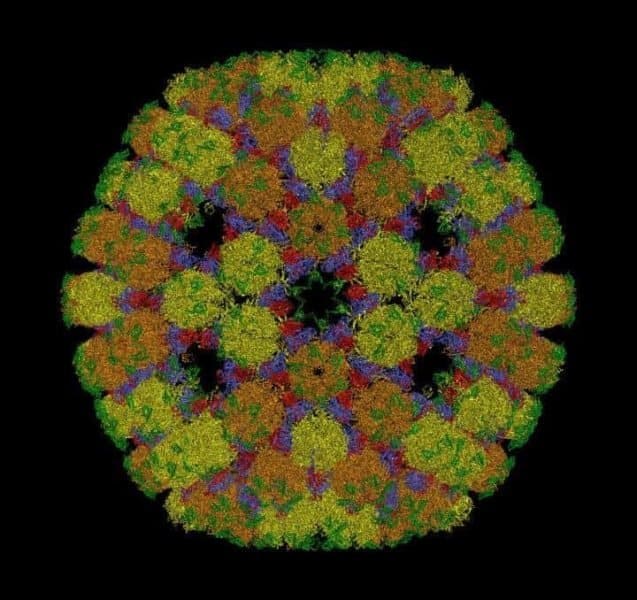
Viruses look like tiny footballs because they package their genetic material into protein containers that adopt polyhedral shapes.
The new theory revolutionises our understanding of how these containers are shaped, solving a scientific mystery that has endured for half a century.
For more than fifty years, scientists have followed the Caspar-Klug theory (CKT) about how the protein containers of viruses are structured. However, improvements in our ability to image viral particles at high resolution have made it apparent that many virus structures do not conform to these blueprints.
Published in the journal Nature Communications, the new theory accurately predicts the positions of proteins in the containers of all icosahedral (or twenty-sided) for the first time. It simultaneously works for viruses that conform to CKT and for those that posed an unresolved problem to that theory.
Professor Reidun Twarock, mathematical biologist at the University of York’s departments of Mathematics and Biology and a member of the York Cross-disciplinary Centre for Systems Analysis, said: “Our study represents a quantum leap forward in the field of structural virology, and closes gaps in our understanding of the structures of many viruses that are ill described by the existing framework.
“This theory will help scientists to analyse the physical properties of viruses, such as their stability, which is important for a better understanding of the mechanism of infection. Such insights can then be exploited for the development of novel anti-viral strategies.
“In particular the structures of larger and more complex viruses that are formed from multiple different components were previously not well understood.
“Our over-arching scheme reveals container architectures with protein numbers that are excluded by the current framework, and thus closes the size gaps in CKT.
“The new blueprints also provide a new perspective on viral evolution, suggesting novel routes in which larger and more complex viruses may have evolved from simple ones at evolutionary timescales.”
Dr Antoni Luque, theoretical biophysicist at San Diego State University and their Viral Information Institute, said: “We can use this discovery to target both the assembly and stability of the capsid, to either prevent the formation of the virus when it infects the host cell, or break it apart after it’s formed. This could facilitate the characterization and identification of antiviral targets for viruses sharing the same icosahedral layout.”
###