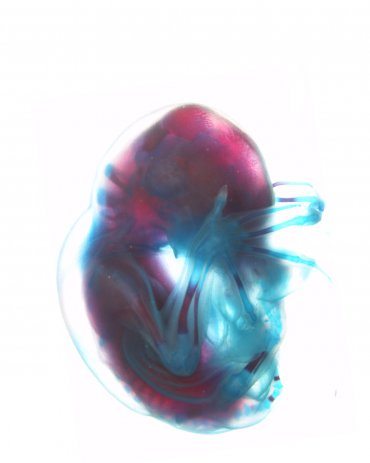
An international team of scientists, including groups from UC San Francisco, Gladstone Institutes, and the University of Cape Town (UCT), South Africa, have for the first time identified genes and gene regulatory elements that are essential in wing development in the Natal long-fingered bat (Miniopterus natalensis), a species widely distributed in eastern and southern Africa.
The new research — presented in two papers published on March 28, one in Nature Genetics and one in PLoS Genetics — revealed regulatory switches that turn bat genes on and off at crucial times during limb development, and has implications for understanding how differences in the size, shape and structure of limbs are generated in mammals in general, including humans, the researchers said.
“This gives us our first detailed picture of the genomics behind bat wing development,” said co-senior investigator Nadav Ahituv, PhD, a UCSF associate professor of bioengineering and therapeutic sciences in the UCSF School of Pharmacy and member of the UCSF Institute for Human Genetics, whose lab also studies the genetics of human limb malformations. “While some attempts have been made to identify the molecular events that led to the evolution of the bat wing, these have been primarily done on a ‘gene by gene’ basis. In contrast, this work lays out a genome-wide blueprint for the causes that led to the development of the bat wing, a key evolutionary innovation that contributed to bats becoming the second most diverse order of mammals.”
Bats are the only mammals capable of powered flight — an innovation that is thought to have occurred about 50 million years ago. Biologists since Charles Darwin have used the structure of the bat wing as an example of both evolutionary novelty — the appearance of a new trait — and vertebrate homology, or shared ancestry between two seemingly different structures — in this case, the wing of the bat and the forelimb of other mammals. But the path of bats’ unique evolution is unclear, noted Nicola Illing, PhD, co-senior investigator in the Department of Molecular and Cell Biology at UCT.
“The fossil record does not show the transition from tree-climbing mammals with short, free digits to ones that have elongated fingers supporting a wing,” Illing said. “Until now, scientists knew very little about how genes are turned on and off during bat embryonic development to transform a mammalian forelimb into a wing.”
Bat Embryos Reveal Thousands of Wing Development Genes
In the Nature Genetics paper, the scientists, including co-lead authors Walter L. Eckalbar, PhD, a postdoctoral fellow in Ahituv’s laboratory at UCSF, and PhD student Stephen Schlebusch of UCT, first sequenced the entire genome of the Natal long-fingered bat. They then performed detailed molecular genomic analysis on bat embryos collected by Illing and her research group at the de Hoop Nature Reserve in South Africa.
The researchers identified over 7,000 genes that are expressed differently in forelimbs compared with hindlimbs at three key stages of bat wing development. They found that many signaling pathways are activated differentially as well, including pathways important in limb formation, digit growth, long bone development and cell death. Also expressed differently are many proteins associated with ribosomes – molecular machines found in all cells that are responsible for protein production during limb development.
“It took bats millions of years to evolve wings,” said Eckalbar. “Our work shows that they did this through thousands of genetic alterations, involving both genes used by all animals during limb development and genes whose usage in limb development may be unique to bats.”
In addition, the scientists found thousands of genetic switches, called enhancers, which regulate the timing and levels of gene expression and show differences in activity between forelimbs and hindlimbs at these key stages of wing development.
“Importantly, this work identified not just which genes are expressed at what stage of growth, but the genetic switches in the genome that are responsible for turning those genes on and off,” Ahituv said.
Researchers Map Bat Wings’ Unique Evolutionary Origins
In the study published in PLoS Genetics, the research team, including co-lead authors Betty M. Booker, PhD, a post-doctoral fellow in Ahituv’s laboratory, and Tara Friedrich, a PhD student at UCSF and Gladstone Institutes, searched for the evolutionary origin of the bat wing.
“We identified genomic sequences that have not changed in most vertebrates, but experienced rapid changes in the common ancestor of today’s bats,” explained Friedrich, a member of the laboratory of co-senior investigator Katherine S. Pollard, PhD, a senior investigator at the Gladstone Institutes, a UCSF professor of epidemiology and biostatistics, and a member of the UCSF Institute for Human Genetics.
The team mapped these so-called “bat accelerated regions” (BARs) onto areas that were predicted to be important switches that turn genes on during limb development, and found 166 BARs with the potential to influence bat wing development.
The researchers tested the effects of five of these BARs in genetically modified mouse embryos and found that all five bat sequences were capable of switching on a reporter gene in the developing mouse forelimb. They noted that one region, BAR116, is located near the HoxD genes, which are known to be involved in limb patterning and skeletal growth.
Previously, Mandy Mason, a PhD student at UCT, had shown that two of the HoxD genes — Hoxd10 and Hoxd11 — are far more active in bat wings compared to bat legs during their embryonic development. Following up these lines of evidence, the researchers showed that the bat BAR116 sequence appears to function as a genetic switch that is active in developing limbs, in particular the forelimbs, while the equivalent mouse sequence did not show any activity.
“Our computational method enabled identification of DNA sequences that changed dramatically during the emergence of bats,” said Pollard. “It is exciting to see that this evolutionary signature pointed us to parts of the mammalian genome that control limb development.”
In addition to unveiling new fundamental details of the evolutionary and developmental origins of powered flight in bats, the new research may provide broader insights into the biological processes that control how mammalian limbs develop in general, Ahituv said. “Importantly, this work will increase our understanding of how alterations in limb development could lead to limb malformations in humans,” he said. “Potentially, it could eventually help contribute to the development of tools and techniques to prevent such malformations.”
Other authors of the Nature Genetics paper are Sierra Nishizaki (PhD student), Nadja Makki, PhD, and Julia E. VanderMeer, PhD, of UCSF; Zoe Gill, MSc, Ash V. Parker (Honours student) and Christina Muswamba-Nday, PhD, of UCT; Kimberly Nevonen of the Oregon National Primate Research Center (ONPRC); Elizabeth Terhune of ONPRC and the University of Colorado, Boulder, CO; and Lucia Carbone, PhD, of ONPRC and Oregon Health and Science University. Jeff D. Wall, PhD, MS, UCSF associate professor of epidemiology and biostatistics and a faculty member at the UCSF Institute for Human Genetics, was a co-senior investigator.
Other authors of the PLoS Genetics paper are Julia E. VanderMeer, PhD, and Jingjing Zhao, PhD, of UCSF, and Malcolm Logan, PhD, of the National Institute for Medical Research and King’s College London, UK.