A new paper recently published in the Journal of Molecular Biology shows how advances in molecular biology and computer science around the world soon may lead to a three-dimensional computer model of a cell, the fundamental unit of life.
According to the authors, the development could herald a new era for biological research, medical science, and human and animal health.
“Cells are the foundation of life,” said Ilya Vakser, professor of computational biology and molecular biosciences and director of the Center for Computational Biology at the University of Kansas, one of the paper’s co-authors. “Recently, there has been tremendous progress in biomolecular modeling and advances at understanding life at the molecular level. Now, the focus is shifting to larger systems — up to the level of the entire cell. We’re trying to capture this emerging milestone development in computational structural biology, which is the tectonic shift from modeling individual biomolecular processes to modeling the entire cell.”
The study, titled “Challenges in structural approaches to cell modeling,” surveys a range of methodologies joining the march toward a simulated whole 3-D cell, including the studies of biological networks, automated construction of 3-D cell models with experimental data, modeling of protein complexes, prediction of protein interactions, thermodynamic and kinetic effects of crowding cellular membrane modeling, and modeling of chromosomes.
“A lot of techniques that are required for this are already available — it’s just a matter of putting them all together in a coherent strategy to address this problem,” Vakser said. “It’s hard because we’re just beginning to understand the principal mechanisms of life at the molecular level — it looks extremely complicated but doable, so we’re moving very fast — not only in our ability to understand how it works at the molecular level but to model it.”
While most of these techniques are being developed separately, the authors say that considered together they represent a push forward that could provide a better basic “understanding of life at the molecular level and lead to important applications to biology and medicine.”
“There are two major benefits,” Vakser said. “One is our fundamental understanding of how a cell works. You can’t claim you understand a phenomenon if you can’t model it. So this gives us insight into basic fundamentals of life at the scale of an entire cell. On the practical side, it will give us an improved grasp of the underlying mechanisms of diseases and also the ability to understand mechanisms of drug action, which will be a tremendous boost to our efforts at drug design. It will help us create better drug candidates, which will potentially shorten the path to new drugs.”
As an example, the KU researcher said a working 3-D molecular cell model could help to replace or augment phases of time-consuming and expensive drug development protocols required today to bring drug therapies from the scientist’s bench to the marketplace.
Vakser said that facets of the research that could lead to a computer-simulated cell are at different levels of refinement.
“We’ve made advances in our ability to model protein interactions,” he said. “The challenge is to put it in context of the cell, which is a densely populated milieu of different proteins and other biomolecular structures. To make the transition from a dilute solution to realistic environment encountered in the cell is probably the greatest challenge we’re facing right now.”
While modeling more complex human cells might be on the agenda soon, Vakser said that for the time being, research efforts will focus on modeling simple single-celled organisms.
“We go for the simplest cell possible. There are small prokaryotic cells, which involve minimalistic set of elements that are much simpler than the bigger and more complicated cells in mammals, including humans,” he said. “We’re trying to cut our teeth on the smallest possible cellular organisms first, then will extrapolate into more complicated cells.”
Along with Vakser, the paper’s authors are Wonpil Im of Lehigh University, Jie Liang of the University of Illinois at Chicago, Arthur Olson of The Scripps Research Institute and Huan-Xiang Zhou of Florida State University.
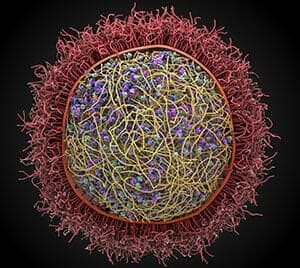
Photo: This image from the Scripps Research Institute shows a preliminary model of mycoplasma mycoides. Modeling by Ludovic Autin and David Goodsell, rendering by Adam Gardner.