One of the most detailed genomic studies of any ecosystem to date has revealed an underground world of stunning microbial diversity, and added dozens of new branches to the tree of life.
The bacterial bonanza comes from scientists who reconstructed the genomes of more than 2,500 microbes from sediment and groundwater samples collected at an aquifer in Colorado. The effort was led by researchers from the Department of Energy’s Lawrence Berkeley National Laboratory (Berkeley Lab) and UC Berkeley. DNA sequencing was performed at the Joint Genome Institute, a DOE Office of Science User Facility.
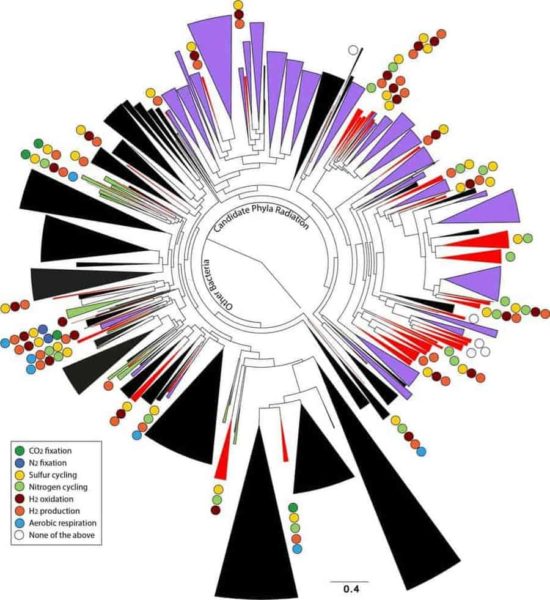
As reported online October 24 in the journal Nature Communications, the scientists netted genomes from 80 percent of all known bacterial phyla, a remarkable degree of biological diversity at one location. They also discovered 47 new phylum-level bacterial groups, naming many of them after influential microbiologists and other scientists. And they learned new insights about how microbial communities work together to drive processes that are critical to the planet’s climate and life everywhere, such as the carbon and nitrogen cycles.
These findings shed light on one of Earth’s most important and least understood realms of life. The subterranean world hosts up to one-fifth of all biomass, but it remains a mystery.
“We didn’t expect to find this incredible microbial diversity. But then again, we know little about the roles of subsurface microbes in biogeochemical processes, and more broadly, we don’t really know what’s down there,” says Jill Banfield, a Senior Faculty Scientist in Berkeley Lab’s Climate & Ecosystem Sciences Division and a UC Berkeley professor in the departments of Earth and Planetary Science, and Environmental Science, Policy, and Management.
UC Berkeley’s Karthik Anantharaman, the first author of the paper, adds, “To better understand what subsurface microbes are up to, our approach is to access their entire genomes. This enabled us to discover a greater interdependency among microbes than we’ve seen before.”
The research is part of a Berkeley Lab-led project called Watershed Function Scientific Focus Area (formerly Sustainable Systems Scientific Focus Area 2.0). The project is developing a predictive understanding of terrestrial environments from the genome to the watershed scale. The field research takes place at a research site near the town of Rifle, Colorado, where for the past several years scientists have conducted experiments designed to stimulate populations of subterranean microbes that are naturally present in very low numbers.
The scientists sent soil and water samples from these experiments to the Joint Genome Institute for terabase-scale metagenomic sequencing. This high-throughput method isolates and purifies DNA from environmental samples, and then sequences one trillion base pairs of DNA at a time. Next, the scientists used bioinformatics tools developed in Banfield’s lab to analyze the data.
Their approach has redrawn the tree of life. Between the 47 new bacterial groups reported in this work, and 35 new groups published last year (also found at the Rifle site), Banfield’s team has doubled the number of known bacterial groups.
With discovery comes naming rights. The scientists named many of the new bacteria groups after Berkeley Lab and UC Berkeley researchers. For example, there’s Candidatus Andersenbacteria, after phylochip inventor Gary Andersen, and there’s Candidatus Doudnabacteria, after CRISPR genome-editing pioneer Jennifer Doudna.
“Berkeley now dominates the tree of life as it does the periodic table,” Banfield says, in a nod to the sixteen elements discovered at Berkeley Lab and UC Berkeley.
Another big outcome is a deeper understanding of the roles subsurface microbes play in globally important carbon, hydrogen, nitrogen, and sulfur cycles. This information will help to better represent these cycles in predictive models such as climate simulations.
The scientists conducted metabolic analyses of 36 percent of the organisms detected in the aquifer system. They focused on a phenomenon called metabolic handoff, which essentially means one microbe’s waste is another microbe’s food. It’s known from lab studies that handoffs are needed in certain reactions, but these interconnected networks are widespread and vastly more complex in the real world.
To understand why it’s important to represent metabolic handoffs as accurately as possible in models, consider nitrate, a groundwater contaminant from fertilizers. Subsurface microbes are the primary driver in reducing nitrate to harmless nitrogen gas. There are four steps in this denitrification process, and the third step creates nitrous oxide—one of the most potent greenhouse gases. The process breaks down if microbes that carry out the fourth step are inactive when a pulse of nitrate enters the system.
“If microbes aren’t there to accept the nitrous oxide handoff, then the greenhouse gas escapes into the atmosphere,” says Anantharaman.
The scientists found the carbon, hydrogen, nitrogen, and sulfur cycles are all driven by metabolic handoffs that require an unexpectedly high degree of interdependence among microbes. The vast majority of microorganisms can’t fully reduce a compound on their own. It takes a team. There are also backup microbes ready to perform a handoff if first-string microbes are unavailable.
“The combination of high microbial diversity and interconnections through metabolic handoffs likely results in high ecosystem resilience,” says Banfield.
Other co-authors of the paper include Berkeley Lab’s Eoin Brodie, Susan Hubbard, Ulas Karaoz, and Kenneth Williams; and UC Berkeley’s Chris Brown, Cindy Castelle, Laura Hug, Alexander Probst, Itai Sharon, Andrea Singh, and Brian Thomas. The research is supported by the Department of Energy’s Office of Science.