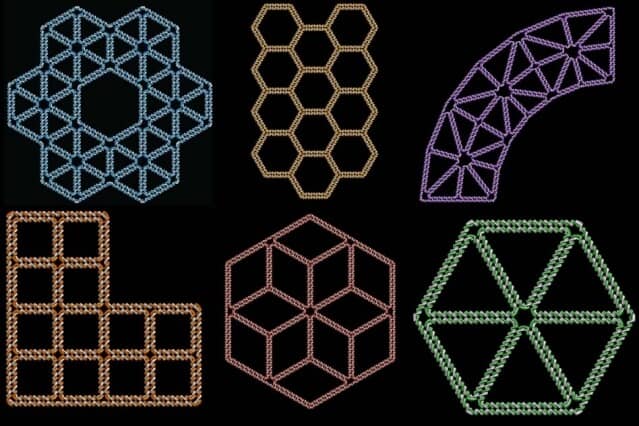
Researchers at MIT and Arizona State University have designed a computer program that allows users to translate any free-form drawing into a two-dimensional, nanoscale structure made of DNA.
Until now, designing such structures has required technical expertise that puts the process out of reach of most people. Using the new program, anyone can create a DNA nanostructure of any shape, for applications in cell biology, photonics, and quantum sensing and computing, among many others.
“What this work does is allow anyone to draw literally any 2-D shape and convert it into DNA origami automatically,” says Mark Bathe, an associate professor of biological engineering at MIT and the senior author of the study.
The researchers published their findings Jan. 2 in Science Advances, and the program, called PERDIX, is available online. The lead authors of the paper are Hyungmin Jun, an MIT postdoc, and Fei Zhang, an assistant research professor at Arizona State University. Other authors are MIT research associate Tyson Shepherd, recent MIT PhD recipient Sakul Ratanalert, ASU assistant research scientist Xiaodong Qi, and ASU professor Hao Yan.
Hyungmin Jun
Automated design
DNA origami, the science of folding DNA into tiny structures, originated in the early 1980s, when Ned Seeman of New York University proposed taking advantage of DNA’s base-pairing abilities to create arbitrary molecular arrangements. In 2006, Paul Rothemund of Caltech created the first scaffolded, two-dimensional DNA structures, by weaving a long single strand of DNA (the scaffold) through the shape such that DNA strands known as “staples” would hybridize to it to help the overall structure maintain its shape.
Others later used a similar approach to create complex three-dimensional DNA structures. However, all of these efforts required complicated manual design to route the scaffold through the entire structure and to generate the sequences of the staple strands. In 2016, Bathe and his colleagues developed a way to automate the process of generating a 3-D polyhedral DNA structure, and in this new study, they set out to automate the design of arbitrary 2-D DNA structures.
To achieve that, they developed a new mathematical approach to the process of routing the single-stranded scaffold through the entire structure to form the correct shape. The resulting computer program can take any free-form drawing and translate it into the DNA sequence to create that shape and into the sequences for the staple strands.
The shape can be sketched in any computer drawing program and then converted into a computer-aided design (CAD) file, which is fed into the DNA design program. “Once you have that file, everything’s automatic, much like printing, but here the ink is DNA,” Bathe says.
After the sequences are generated, the user can order them to easily fabricate the specified shape. In this paper, the researchers created shapes in which all of the edges consist of two duplexes of DNA, but they also have a working program that can utilize six duplexes per edge, which are more rigid. The corresponding software tool for 3-D polyhedra, called TALOS, is available online and will be published soon in the journal ACS Nano. The shapes, which range from 10 to 100 nanometers in size, can remain stable for weeks or months, suspended in a buffer solution.
“The fact that we can design and fabricate these in a very simple way helps to solve a major bottleneck in our field,” Bathe says. “Now the field can transition toward much broader groups of people in industry and academia being able to functionalize DNA structures and deploy them for diverse applications.”
Nanoscale patterns
Because the researchers have such precise control over the structure of the synthetic DNA particles, they can attach a variety of other molecules at specific locations. This could be useful for templating antigens in nanoscale patterns to shed light on how immune cells recognize and are activated by specific arrangements of antigens found on viruses and bacteria.
“How nanoscale patterns of antigens are recognized by immune cells is a very poorly understood area of immunology,” Bathe says. “Attaching antigens to structured DNA surfaces to display them in organized patterns is a powerful way to probe that biology.”
Another key application is designing light-harvesting circuits that mimic the photosynthetic complexes found in plants. To achieve that, the researchers are attaching light-sensitive dyes known as chromophores to DNA scaffolds. In addition to harvesting light, such circuits could also be used to perform quantum sensing and rudimentary computations. If successful, these would be the first quantum computing circuits that can operate at room temperature, Bathe says.
Bathe and three other MIT faculty members — Gabriela Schlau-Cohen, Adam Willard, and Dirk Englund — recently received a National Science Foundation grant to pursue this quantum sensing and computing project.
Other possible applications for the DNA structures include using them to help organize macromolecular protein assemblies found in cells, so that they can be more easily imaged with high-resolution cryo-electron-microscopy. MIT’s new MIT.nano facility has two such microscopes, which can be used to reveal the fine details of tiny structures.
The research was funded by the National Science Foundation, the Office of Naval Research, the Army Research Office, and the National Institute of Allergy and Infectious Disease.