Computer models are an important tool for studying how the brain makes and stores memories and other types of complex information. But creating such models is a tricky business. Somehow, a symphony of signals – both biochemical and electrical – and a tangle of connections between neurons and other cell types creates the hardware for memories to take hold. Yet because neuroscientists don’t fully understand the underlying biology of the brain, encoding the process into a computer model in order to study it further has been a challenge.
Now, researchers at the Okinawa Institute of Science and Technology (OIST) have altered a commonly used computer model of memory called a Hopfield network in a way that improves performance by taking inspiration from biology. They found that not only does the new network better reflect how neurons and other cells wire up in the brain, it can also hold dramatically more memories.
The complexity added to the network is what makes it more realistic, says Thomas Burns, a PhD student in the group of Professor Tomoki Fukai, who heads OIST’s Neural Coding and Brain Computing Unit. “Why would biology have all this complexity? Memory capacity might be a reason,” Mr. Burns says.
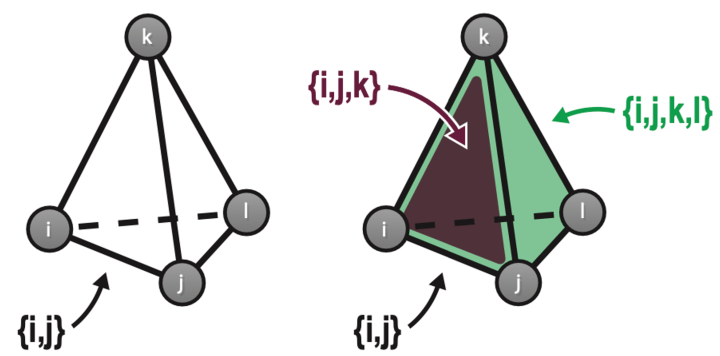
Hopfield networks store memories as patterns of weighted connections between different neurons in the system. The network is “trained” to encode these patterns, then researchers can test its memory of them by presenting a series of blurry or incomplete patterns and seeing if the network can recognize them as one it already knows. In classical Hopfield networks, however, neurons in the model reciprocally connect to other neurons in the network to form a series of what are called “pairwise” connections. Pairwise connections represent how two neurons connect at a synapse, a connection point between two neurons in the brain. But in reality, neurons have intricate branched structures called dendrites that provide multiple points for connection, so the brain relies on a much more complex arrangement of synapses to get its cognitive jobs done. Additionally, connections between neurons are modulated by other cell types called astrocytes.
“It’s simply not realistic that only pairwise connections between neurons exist in the brain,” explains Mr. Burns. He created a modified Hopfield network in which not just pairs of neurons but sets of three, four, or more neurons could link up too, such as might occur in the brain through astrocytes and dendritic trees. Although the new network allowed these so-called “set-wise” connections, over all it contained the same total number of connections as before. The researchers found that a network containing a mix of both pairwise and set-wise connections performed best and retained the highest number of memories. They estimate it works more than doubly as well as a traditional Hopfield network. “It turns out you actually need a combination of features in some balance,” says Mr. Burns. “You should have individual synapses, but you should also have some dendritic trees and some astrocytes.”
Hopfield networks are important for modeling brain processes, but they have powerful other uses too. For example, very similar types of networks called Transformers underlie AI-based language tools such as ChatGPT, so the improvements Mr. Burns and Professor Fukai have identified may also make such tools more robust.
Mr. Burns and his colleagues plan to continue working with their modified Hopfield networks to make them still more powerful . For example, in the brain the strengths of connections between neurons are not normally the same in both directions, so Mr. Burns wonders if this feature of asymmetry might also improve the network’s performance. Additionally, he would like to explore ways of making the network’s memories interact with each other, the way they do in the human brain. “Our memories are multifaceted and vast,” says Mr. Burns. “We still have a lot to uncover.”
The study is published as a conference paper, entitled, “Simplicial Hopfield networks,” in International Conference on Learning Representations.