Duke researchers have devised a computerized method to autonomously and quickly diagnose malaria with clinically relevant accuracy — a crucial step to successfully treating the disease and halting its spread.
In 2015 alone, malaria infected 214 million people worldwide, killing an estimated 438,000.
While Western medicine can spot malaria with near-perfect accuracy, it can be difficult to diagnose in resource-limited areas where infection rates are highest.
Malaria’s symptoms can look like many other diseases, and there are simply not enough well-trained field workers and functioning microscopes to keep pace with the parasite. While rapid diagnostic tests do exist, it is expensive to continuously purchase new tests. These tests also cannot tell how severe the infection is by tallying the number of infected cells, which is important for managing a patient’s recovery.
In a new study, engineers from Duke University report a method that uses computer ‘deep learning’ and light-based, holographic scans to spot malaria-infected cells from a simple, untouched blood sample without any help from a human. The innovation could form the basis of a fast, reliable test that could be given by most anyone, anywhere in the field, which would be invaluable in the $2.7 billion-per-year global fight against the disease.
The results were published online Sept. 16 in the journal PLOS ONE.
“With this technique, the path is there to be able to process thousands of cells per minute,” said Adam Wax, professor of biomedical engineering at Duke. “That’s a huge improvement to the 40 minutes it currently takes a field technician to stain, prepare and read a slide to personally look for infection.”
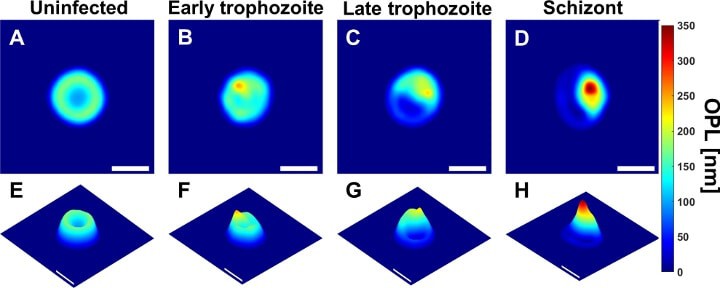
The new technique is based on a technology called quantitative phase spectroscopy. As a laser sweeps through the visible spectrum of light, sensors capture how each discrete light frequency interacts with a sample of blood. The resulting data captures a holographic image that provides a wide array of valuable information that can indicate a malarial infection.
“We identified 23 parameters that are statistically significant for spotting malaria,” said Han Sang Park, a doctoral student in Wax’s laboratory and first author on the paper. For example, as the disease progresses, red blood cells decrease in volume, lose hemoglobin and deform as the parasite within grows larger. This affects features such as cell volume, perimeter, shape and center of mass.
“However, none of the parameters were reliable more than 90 percent of the time on their own, so we decided to use them all,” said Park.
“To be adopted, any new diagnostic device has to be just as reliable as a trained field worker with a microscope,” said Wax. “Otherwise, even with a 90 percent success rate, you’d still miss more than 20 million cases a year.”
To get a more accurate reading, Wax and Park turned to deep learning — a method by which computers teach themselves how to distinguish between different objects. By feeding data on more than 1,000 healthy and diseased cells into a computer, the deep learning program determined which sets of measurements at which thresholds most clearly distinguished healthy from diseased cells.
When they put the resulting algorithm to the test with hundreds of cells, it was able to correctly spot malaria 97 to 100 percent of the time — a number the researchers believe will increase as more cells are used to train the program. Because the technique breaks data-rich holograms down to just 23 numbers, tests can be easily transmitted in bulk, which is important for locations that often do not have reliable, fast internet connections, and that, in turn, could eliminate the need for each location to have its own computer for processing.
Wax and Park are now looking to develop the technology into a diagnostic device through a startup company called M2 Photonics Innovations. They hope to show that a device based on this technology would be accurate and cost-efficient enough to be useful in the field. Wax has also received funding to begin exploring the use of the technique for spotting cancerous cells in blood samples.
This work was supported by the National Science Foundation (IIP-1346349), the National Institutes of Health (R01-CA167421), the World Anti-Doping Agency, and the Burroughs Wellcome Fund.
“Automated detection of P. falciparum using machine learning algorithms with quantitative phase images of unstained cells.” Han Sang Park, Matthew T. Rinehart, Katelyn A. Walzer, Jen-Tsan Ashley Chi, Adam Wax. PLOS ONE, 2016. DOI: 10.1371/journal.pone.0163045