With an unparalleled collection of rare biological samples and expertise in profiling RNA sequences, Duke University physicians and engineers are playing a vital role in a major U.S. Defense Advanced Research Projects Agency (DARPA) initiative to rapidly screen individuals for exposure to materials associated with weapons of mass destruction (WMD).
The $38.8-million project, led by Arizona State University, joins a $27.8 project led by Mt. Sinai as one of two national consortiums funded by DARPA’s new Epigenetic CHaracterization and Observation (ECHO) program.
The end goal of ECHO is to build a field-deployable device that can test small biological samples—such as a drop of blood or nasal swab—for evidence of epigenetic “fingerprints” that reveal a detailed history of that individual’s exposure to dangerous materials or pathogens.
Although an individual’s DNA remains the same over their lifetime, their environment can affect how his or her genes are expressed, and these changes leave a lasting imprint on their epigenome—even long after the triggering event has past. According to DARPA, technology that could rapidly read these epigenetic signatures could speed up diagnosis and treatment for troops who may have been exposed to threat agents or who may be suffering from infections.
In a project dubbed MEMENTO—Mapping Epigenetic Memory of Exposure to Observe—Duke has been awarded $6.2 million over the next four years to help make this idea a reality.
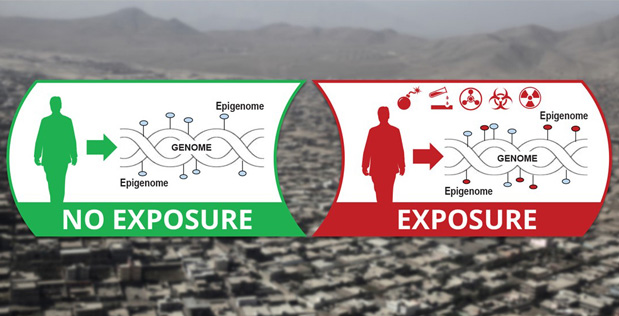
DARPA’s new Epigenetic CHaracterization and Observation (ECHO) program aims to build a field-deployable platform technology that quickly reads someone’s epigenome and identifies signatures that indicate whether that person has ever—in his or her lifetime—been exposed to materials that could be associated with weapons of mass destruction (WMD)
Led by co-principal investigators Xiling Shen, the Hawkins Family Associate Professor of Biomedical Engineering at Duke, and Christopher Woods, professor of medicine and global health at Duke and chief of the infectious diseases section at the Durham VA Medical Center, the project draws on a treasure trove of rare biological samples to help researchers across the ECHO program discover specific epigenetic signatures associated with various exposures.Duke Global Health Institute researchers will also collect a number of new sample cohorts for use in the DARPA projects to study how epigenetic markers persist over time. Gayani Tillekeratne, assistant professor of medicine and global health, will collect samples from subjects with confirmed dengue virus infection in southern Sri Lanka, where Dharshan de Silva, director and senior scientist at Genetech Research Institute, will pursue a similar cohort of subjects with Burkholderia pseudomallei, the cause of melioidosis. Woods will work with colleagues from Tulane in Sierra Leone to gather specimens from survivors of the Ebola-like hemorrhagic disease Lassa fever and with UCLA investigators in the Democratic Republic of Congo to gather samples from survivors of Ebola outbreaks, including from the very first in 1976.
The resources also include a 20-year historical library of Methicillin-resistant Staphylococcus aureus (MRSA) samples collected by Duke School of Medicine professor Vance Fowler. Lisa Satterwhite, assistant research professor of civil and environmental engineering at Duke, will provide samples from an ongoing program studying the effects of farmworkers’ exposures to organophosphates, which are used in insecticides and are precursors to nerve agents used in chemical warfare.
“Our clinical team has an overarching responsibility to design studies and provide samples to both DARPA projects,” said Woods. “It’s a novel, exciting program that is looking at epigenetic signatures in both real-time exposures as well as their evolution over time. And while there’s a clear defense angle to the work, there’s also a practical clinical understanding that will be beneficial as well.”
On the engineering side, Duke researchers will use their expertise in three challenging RNA and epigenetic analysis techniques and machine learning to identify regions of DNA called enhancers activated by exposure to these diseases.
While only about one percent of the human genome encodes directions for the making of proteins, buried within the remaining “dark genome” are regions called enhancers that can turn on the protein-makers on.
“Enhancers are these control knobs that remotely dial up or down the activity genes,” said Shen. “Our hypothesis is that these enhancers are most likely where the epigenetic memories of exposures to harmful agents are kept.”
To get a full picture of DNA activity, Duke researchers are combining three separate techniques. In Shen’s laboratory, research assistant professor Shengli Ding and PhD student Nick Giroux will perform total RNA sequencing to discover which segments of the dark genome are active and microRNA sequencing to reveal molecules released to stop other RNA from delivering their instructions. Purushothama Rao Tata, assistant professor of cell biology, and Yoshihiko Kobayashi, postdoctoral associate of cell biology, will perform a cutting-edge technique named Mint-ChIP to identify sections of DNA packed away by proteins called histones, allowing researchers to isolate them and remove the proteins to discover which segments of DNA they were activating or silencing.
“We have to use all three of these profiling techniques to infer their activity,” said Shen. “By pursuing a multi-omics approach that compiles a wide variety of profiling techniques, we can develop a comprehensive signature that tells us if a person has had a harmful exposure and to what specific agent.”
Other partners in the collaboration will also be analyzing these rare exposure samples, but through different methods focused on other alterations to the epigenome and the DNA itself. They will also work with exposures to different types of WMDs, such as radiation, chemical weapons and explosives.
Once signatures are discovered, partner institutions will begin developing a compact, lightweight device capable of scanning biological fluids to detect them.
“The human body registers exposures and logs them in the epigenome,” explained Eric Van Gieson, the ECHO program manager, in a release announcing the program. “We are just beginning to understand this rich biographical record that we carry around with us. We hope that with the capabilities developed within ECHO, someone in the field will immediately know if a suspected adversary has handled or been exposed to threat agents. The same technology could also serve as a diagnostic tool for our own troops, to diagnose infectious disease or reveal exposure to threat agents, so that medical countermeasures can be applied in time to make a difference.”
For more information about the ECHO program, read the full announcement from Arizona State University.