Under a microscope, the first few hours of every multicellular organism’s life seem incongruously chaotic. After fertilization, a once tranquil single-celled egg divides again and again, quickly becoming a visually tumultuous mosh pit of cells jockeying for position inside the rapidly growing embryo.
Yet, amid this apparent pandemonium, cells begin to self-organize. Soon, spatial patterns emerge, serving as the foundation for the construction of tissues, organs, and elaborate anatomical structures from brains to toes and everything in between. For decades, scientists have intensively studied this process, called morphogenesis, but it remains in many ways enigmatic.
Now, researchers at Harvard Medical School and the Institute of Science and Technology (IST) Austria have discovered a key control mechanism that cells use to self-organize in early embryonic development. The findings, published in Science on Oct. 2, shed light on a process fundamental to multicellular life and open new avenues for improved tissue and organ engineering strategies.
Studying spinal cord formation in zebrafish embryos, a team co-led by Sean Megason, professor of systems biology in the Blavatnik Institute at Harvard Medical School (HMS), revealed that different cell types express unique combinations of adhesion molecules in order to self-sort during morphogenesis. These “adhesion codes” determine which cells prefer to stay connected, and how strongly they do so, even as widespread cellular rearrangements occur in the developing embryo.
“My lab’s goal is to understand the basic design principles of biological form,” said Megason, co-corresponding author on the study. “Our findings represent a new way of approaching the question of morphogenesis, which is one of the oldest and most important in embryology. We see this as the tip of the iceberg for such efforts.”
Insights into how cells self-organize in early development could also aid efforts to engineer tissues and organs for clinical uses such as transplantation, the authors said.
“Constructing artificial tissues for research or medical applications is a critically important goal, but currently one of the biggest problems is inconsistency,” said lead study author Tony Tsai, research fellow in systems biology in the Blavatnik Institute at Harvard Medical School. “There is a clear lesson to learn from understanding and reverse engineering how cells in a developing embryo are able to build the components of an organism in such a robust and reproducible way.”
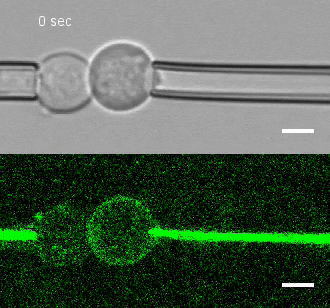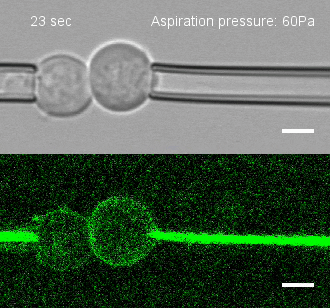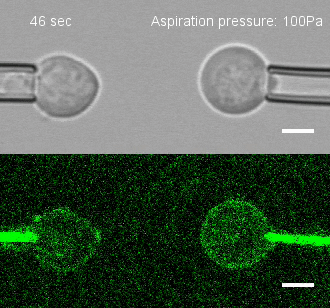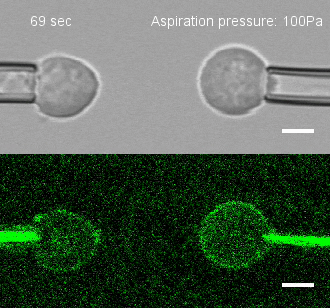
Tug of war
Spearheaded by Tsai and in collaboration with Carl-Philipp Heisenberg and colleagues at IST Austria, the research team first looked at one of the most well-established frameworks for morphogenesis, the French flag model.
In this model, morphogens are released from localized sources in the embryo, exposing nearby cells to higher levels of the signaling molecule than cells farther away. The amount of morphogen a cell is exposed to activates different cellular programs, particularly those that determine cell fate. Concentration gradients of morphogens therefore “paint” patterns onto groups of cells, evocative of the distinct color bands of the French flag.
This model has limitations, however. Previous studies from the Megason lab used live-cell imaging and single-cell tracking in whole zebrafish embryos to show that morphogen signals can be noisy and imprecise, particularly at the boundaries of the “flag.” In addition, cells in a developing embryo are constantly dividing and in motion, which can scramble the morphogen signal. This results in an initial mixed patterning of cell types.
Nevertheless, cells self-sort into precise patterns, even with a noisy start, and in the current study, the team set out to understand how. They focused on a hypothesis proposed over 50 years ago, known as differential adhesion. This model suggests that cells adhere to certain other cell types, self-sorting in a way similar to how oil and vinegar separates over time. But there was little evidence that this plays a role in patterning.
To investigate, Megason, Tsai and colleagues developed a method to measure the force by which cells adhere to one another. They placed two individual cells together and then pulled on each cell with precisely controlled suction pressure from two micropipettes. This allowed the researchers to measure the precise amount of force needed to pull the cells apart. By analyzing three cells at once, they could also establish adhesion preferences.
The team used this technique to study the patterning of three different types of neural progenitor cells involved in building the nascent spinal cord in zebrafish embryos.
The experiments revealed that cells of a similar type strongly and preferentially adhered to one another. To identify the relevant adhesion molecule-encoding genes, the researchers analyzed the gene expression profile of each cell type using RNA sequencing. They then used CRISPR-Cas9 to block the expression of candidate genes, one at a time. If pattern formation became disrupted, they applied the pulling assay to see how much the molecule contributed to adhesion.
Adhesion code
Three genes — N-cadherin, cadherin 11 and protocadherin 19 — emerged as essential for normal patterning. The expression of different combinations and different levels of these genes was responsible for differences in adhesion preference, representing what the team dubbed an adhesion code. This code was unique to each of the cell types and determined which other cells each cell type stays connected to during morphogenesis.
“All three adhesion molecules we looked at are expressed in different amounts in each cell type,” Tsai said. “Cells use this code to preferentially adhere to cells of their own type, which is what allows different cell types to separate during pattern formation. But cells also maintain some level of adhesion with other cell types since they have to collaborate to form tissues. By piecing together these local interaction rules, we can illuminate the global picture.”
Because the adhesion code is cell-type specific, the researchers hypothesized that it is likely controlled by the same processes that determine cell fate — namely, morphogen signaling. They looked at how perturbations to one the most well-known morphogens, Sonic hedgehog (Shh), affected cell type and corresponding adhesion-molecule gene expression.
The analyses revealed that both cell type and adhesion-molecule gene expression were highly correlated, both in level and spatial position. This held true across the entire nascent spinal cord, where patterns of gene expression for cell type and adhesion molecule changed together in response to differences in Shh activity.
“What we found is that this morphogen not only controls cell fate, it controls cell adhesion,” Megason said. “The French flag model gives a rough sketch, and differential adhesion then forms the precise pattern. Combining these different strategies appears to be how cells build patterns in 3D space and time as the embryo is forming.”
The researchers are now further investigating the interplay between morphogen signaling and adhesion in developing embryos. The current study looked at only three different cell types, and there are many other adhesion-molecule candidates and morphogens that remain to be analyzed, the authors said. In addition, the details of how morphogens control both cell type and adhesion molecule expression remain unclear.
Better understanding these processes could help scientists uncover and reverse engineer the fundamental mechanisms by which a single-celled egg constructs a whole organism, the authors said. This could have profound implications in biotechnology, particularly for efforts to build artificial tissues and organs for transplantation or for testing new drug candidates.
“The issue with tissue engineering right now is that we just don’t know what the underlying science is,” Megason said. “If you want to build a little bridge over a stream, maybe you could do that without understanding physics. But if you wanted to build a big suspension bridge, you need to know a lot about the underlying physics. Our goal is to figure out what those rules are for the embryo.”
Additional authors on the study include Mateusz Sikora, Peng Xia, Tugba Colak-Champollion and Holger Knaut.
This work was supported by the National Institutes of Health (grants R01GM107733, R01NS102322), a K99 fellowship from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (1K99HD092623), the Damon Runyon Cancer Foundation, the Company of Biologists, the Burroughs Wellcome Fund and the European Research Council.


