Using an ordinary light microscope, MIT engineers have devised a technique for imaging biological samples with accuracy at the scale of 10 nanometers — which should enable them to image viruses and potentially even single biomolecules, the researchers say.
The new technique builds on expansion microscopy, an approach that involves embedding biological samples in a hydrogel and then expanding them before imaging them with a microscope. For the latest version of the technique, the researchers developed a new type of hydrogel that maintains a more uniform configuration, allowing for greater accuracy in imaging tiny structures.
This degree of accuracy could open the door to studying the basic molecular interactions that make life possible, says Edward Boyden, the Y. Eva Tan Professor in Neurotechnology, a professor of biological engineering and brain and cognitive sciences at MIT, and a member of MIT’s McGovern Institute for Brain Research and Koch Institute for Integrative Cancer Research.
“If you could see individual molecules and identify what kind they are, with single-digit-nanometer accuracy, then you might be able to actually look at the structure of life. And structure, as a century of modern biology has told us, governs function,” says Boyden, who is the senior author of the new study.
The lead authors of the paper, which appears today in Nature Nanotechnology, are MIT Research Scientist Ruixuan Gao and Chih-Chieh “Jay” Yu PhD ’20. Other authors include Linyi Gao PhD ’20; former MIT postdoc Kiryl Piatkevich; Rachael Neve, director of the Gene Technology Core at Massachusetts General Hospital; James Munro, an associate professor of microbiology and physiological systems at University of Massachusetts Medical School; and Srigokul Upadhyayula, a former assistant professor of pediatrics at Harvard Medical School and an assistant professor in residence of cell and developmental biology at the University of California at Berkeley.
Low cost, high resolution
Many labs around the world have begun using expansion microscopy since Boyden’s lab first introduced it in 2015. With this technique, researchers physically enlarge their samples about fourfold in linear dimension before imaging them, allowing them to generate high-resolution images without expensive equipment. Boyden’s lab has also developed methods for labeling proteins, RNA, and other molecules in a sample so that they can be imaged after expansion.
“Hundreds of groups are doing expansion microscopy. There’s clearly pent-up demand for an easy, inexpensive method of nanoimaging,” Boyden says. “Now the question is, how good can we get? Can we get down to single-molecule accuracy? Because in the end, you want to reach a resolution that gets down to the fundamental building blocks of life.”
Other techniques such as electron microscopy and super-resolution imaging offer high resolution, but the equipment required is expensive and not widely accessible. Expansion microscopy, however, enables high-resolution imaging with an ordinary light microscope.
In a 2017 paper, Boyden’s lab demonstrated resolution of around 20 nanometers, using a process in which samples were expanded twice before imaging. This approach, as well as the earlier versions of expansion microscopy, relies on an absorbent polymer made from sodium polyacrylate, assembled using a method called free radical synthesis. These gels swell when exposed to water; however, one limitation of these gels is that they are not completely uniform in structure or density. This irregularity leads to small distortions in the shape of the sample when it’s expanded, limiting the accuracy that can be achieved.
To overcome this, the researchers developed a new gel called tetra-gel, which forms a more predictable structure. By combining tetrahedral PEG molecules with tetrahedral sodium polyacrylates, the researchers were able to create a lattice-like structure that is much more uniform than the free-radical synthesized sodium polyacrylate hydrogels they previously used.
The researchers demonstrated the accuracy of this approach by using it to expand particles of herpes simplex virus type 1 (HSV-1), which have a distinctive spherical shape. After expanding the virus particles, the researchers compared the shapes to the shapes obtained by electron microscopy and found that the distortion was lower than that seen with previous versions of expansion microscopy, allowing them to achieve an accuracy of about 10 nanometers.
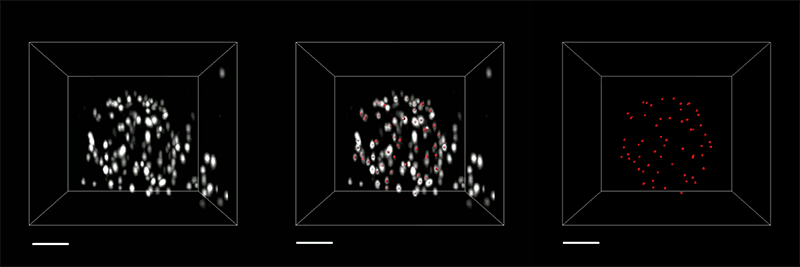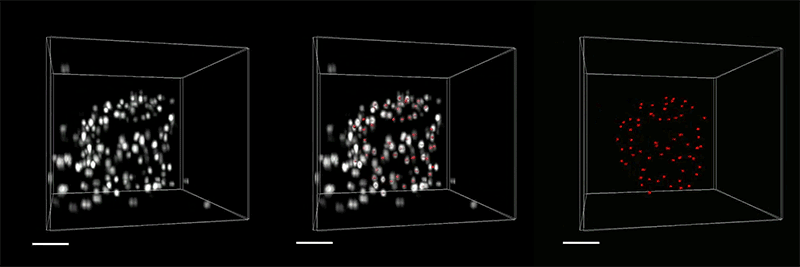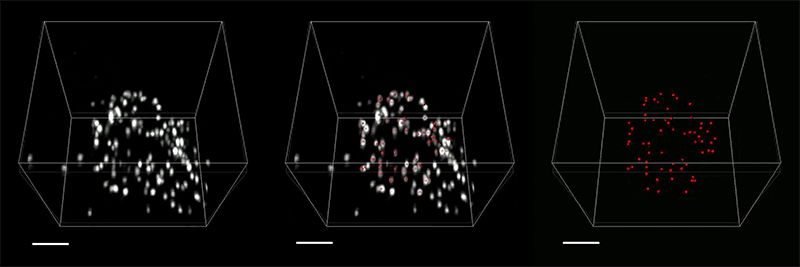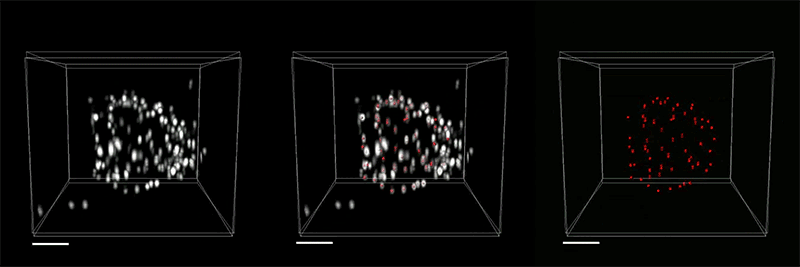
“We can look at how the arrangements of these proteins change as they are expanded and evaluate how close they are to the spherical shape. That’s how we validated it and determined how faithfully we can preserve the nanostructure of the shapes and the relative spatial arrangements of these molecules,” Ruixuan Gao says.
Single molecules
The researchers also used their new hydrogel to expand cells, including human kidney cells and mouse brain cells. They are now working on ways to improve the accuracy to the point where they can image individual molecules within such cells. One limitation on this degree of accuracy is the size of the antibodies used to label molecules in the cell, which are about 10 to 20 nanometers long. To image individual molecules, the researchers would likely need to create smaller labels or to add the labels after expansion was complete.
They are also exploring whether other types of polymers, or modified versions of the tetra-gel polymer, could help them realize greater accuracy.
If they can achieve accuracy down to single molecules, many new frontiers could be explored, Boyden says. For example, scientists could glimpse how different molecules interact with each other, which could shed light on cell signaling pathways, immune response activation, synaptic communication, drug-target interactions, and many other biological phenomena.
“We’d love to look at regions of a cell, like the synapse between two neurons, or other molecules involved in cell-cell signaling, and to figure out how all the parts talk to each other,” he says. “How do they work together and how do they go wrong in diseases?”
The research was funded by Lisa Yang, John Doerr, Open Philanthropy, the National Institutes of Health, the Howard Hughes Medical Institute Simons Faculty Scholars Program, the Intelligence Advanced Research Projects Activity, the U.S. Army Research Laboratory, the US-Israel Binational Science Foundation, the National Science Foundation, the Friends of the McGovern Fellowship, and the Fellows program of the Image and Data Analysis Core at Harvard Medical School.


