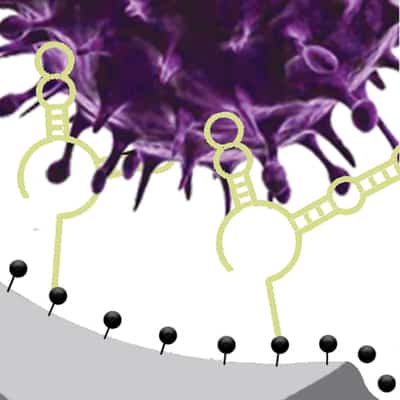
A new sensor can detect not only whether a virus is present, but whether it’s infectious – an important distinction for containing viral spread.
Researchers at the University of Illinois Urbana-Champaign and collaborators developed the sensor, which integrates specially designed DNA fragments and nanopore sensing, to target and detect infectious viruses in minutes without the need to pre-treat samples. They demonstrated the sensor’s power with two key viruses that cause infections worldwide: the human adenovirus and the virus that causes COVID-19.
Yi Lu, a professor emeritus of chemistry, and Benito Marinas, a professor of civil and environmental engineering, co-led the work with University of Illinois Chicago professor Lijun Rong; professor Omar Azzaroni, of the National University of La Plata in Argentina; and María Eugenia Toimil-Molares, of the GSI Helmholtz Centre for Heavy Ion Research in Germany. They reported their findings in the journal Science Advances.
“The infectivity status is very important information that can tell us if patients are contagious or if an environmental disinfection method works,” said Ana Peinetti, the first author of the study, who performed the work while a postdoctoral researcher at Illinois. She now leads a research group at the University of Buenos Aires in Argentina. “Our sensor combines two key components: highly specific DNA molecules and highly sensitive nanopore technology. We developed these highly specific DNA molecules, named aptamers, that not only recognize viruses but also can differentiate the infectivity status of the virus.”
The “gold standard” of viral detection, PCR tests detect viral genetic material but cannot distinguish whether a sample is infectious or determine whether a person is contagious. This can make it more difficult to track and contain viral outbreaks, the researchers said.
“With the virus that causes COVID-19, it has been shown that the level of viral RNA has minimal correlation with the virus’s infectivity. In the early stage when a person is infected, the viral RNA is low and difficult to detect, but the person is highly contagious,” Lu said. “When a person is recovered and not infectious, the viral RNA level can be very high. Antigen tests follow a similar pattern, though even later than viral RNA. Therefore, viral RNA and antigen tests are both poor in informing whether a virus is infectious or not. It may result in delayed treatment or quarantine, or premature release of those who may still be contagious.”
Tests that detect infectious viruses, called plaque assays, exist but require special preparation and days of incubation to render results. The new sensing method can yield results in 30 minutes to two hours, the researchers report, and since it requires no pre-treatment of the sample, it can be used on viruses that will not grow in the lab.
Being able to distinguish infectious from noninfectious viruses and to detect small amounts from untreated samples that may contain other contaminants is important not only for rapid diagnosis of patients who are in the early stage of infection or who are still contagious after treatment, but for environmental monitoring as well, Marinas said.
“We chose human adenovirus to demonstrate our sensor because it is an emerging waterborne viral pathogen of concern in the United States and throughout the world,” Marinas said. “The capability to detect infectious adenovirus in the presence of viruses rendered noninfectious by water disinfectants, and other potentially interfering background substances in wastewaters and contaminated natural waters, provides an unprecedented novel approach. We see potential for such technology to provide more robust protection of environmental and public health.”
The sensing technique could be applied to other viruses, the researchers say, by tweaking the DNA to target different pathogens. The DNA aptamers used in the sensor can be readily produced with widely available DNA synthesizers, similarly to the RNA probes produced for PCR tests. Nanopore sensors are also commercially available, making the sensing technique readily scalable, said Lu, now a professor at the University of Texas, Austin.
The researchers are working to further improve the sensors’ sensitivity and selectivity, and are integrating their DNA aptamers with other detection methods, such as color-changing dipsticks or sensors to work with smartphones, to eliminate the need for special equipment. With the ability to distinguish noninfectious from infectious viruses, the researchers said they hope their technology could also aid in understanding mechanisms of infection.
“In addition, the aptamer technology could be further developed into multichannel platforms for detecting other emerging waterborne viral pathogens of public and environmental health concern, such as norovirus and enteroviruses, or for variants of the virus that causes COVID-19,” Marinas said.
This work was supported by a RAPID grant from the National Science Foundation and a seed grant from the Institute for Sustainability, Energy, and Environment at Illinois and the Illinois-JITRI Institute. Peinetti was supported by the PEW Latin American Fellowship.