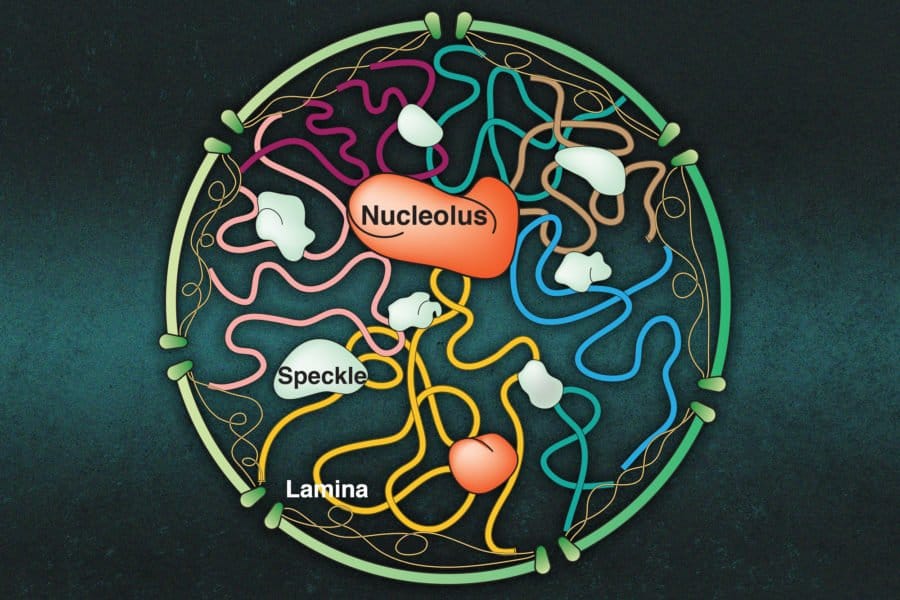
A cell stores all of its genetic material in its nucleus, in the form of chromosomes, but that’s not all that’s tucked away in there. The nucleus is also home to small bodies called nucleoli — clusters of proteins and RNA that help build ribosomes.
Using computer simulations, MIT chemists have now discovered how these bodies interact with chromosomes in the nucleus, and how those interactions help the nucleoli exist as stable droplets within the nucleus.
Their findings also suggest that chromatin-nuclear body interactions lead the genome to take on a gel-like structure, which helps to promote stable interactions between the genome and transcription machineries. These interactions help control gene expression.
“This model has inspired us to think that the genome may have gel-like features that could help the system encode important contacts and help further translate those contacts into functional outputs,” says Bin Zhang, the Pfizer-Laubach Career Development Associate Professor of Chemistry at MIT, an associate member of the Broad Institute of Harvard and MIT, and the senior author of the study.
MIT graduate student Yifeng Qi is the lead author of the paper, which appears today in Nature Communications.
Modeling droplets
Much of Zhang’s research focuses on modeling the three-dimensional structure of the genome and analyzing how that structure influences gene regulation.
In the new study, he wanted to extend his modeling to include the nucleoli. These small bodies, which break down at the beginning of cell division and then re-form later in the process, consist of more than a thousand different molecules of RNA and proteins. One of the key functions of the nucleoli is to produce ribosomal RNA, a component of ribosomes.
Recent studies have suggested that nucleoli exist as multiple liquid droplets. This was puzzling because under normal conditions, multiple droplets should eventually fuse together into one large droplet, to minimize the surface tension of the system, Zhang says.
“That’s where the problem gets interesting, because in the nucleus, somehow those multiple droplets can remain stable across an entire cell cycle, over about 24 hours,” he says.
To explore this phenomenon, Zhang and Qi used a technique called molecular dynamics simulation, which can model how a molecular system changes over time. At the beginning of the simulation, the proteins and RNA that make up the nucleoli are randomly distributed throughout the nucleus, and the simulation tracks how they gradually form small droplets.
In their simulation, the researchers also included chromatin, the substance that makes up chromosomes and incudes proteins as well as DNA. Using data from previous experiments that analyzed the structure of chromosomes, the MIT team calculated the interaction energy of individual chromosomes, which allowed them to provide realistic representations of 3D genome structures.
Using this model, the researchers were able to observe how nucleoli droplets form. They found that if they modeled the nucleolar components on their own, with no chromatin, they would eventually fuse into one large droplet, as expected. However, once chromatin was introduced into the model, the researchers found that the nucleoli formed multiple droplets, just as they do in living cells.
The researchers also discovered why that happens: The nucleoli droplets become tethered to certain regions of the chromatin, and once that happens, the chromatin acts as a drag that prevents the nucleoli from fusing to each other.
“Those forces essentially arrest the system into those small droplets and hinder them from fusing together,” Zhang says. “Our study is the first to highlight the importance of this chromatin network that could significantly slow down the fusion and arrest the system in its droplet state.”
Gene control
The nucleoli are not the only small structures found in the nucleus — others include nuclear speckles and the nuclear lamina, an envelope that surrounds the genome and can bind to chromatin. Zhang’s group is now working on modeling the contributions of these nuclear structures, and their initial findings suggest that they help to give the genome more gel-like properties, Zhang says.
“This coupling that we have observed between chromatin and nuclear bodies is not specific to the nucleoli. It’s general to other nuclear bodies as well,” he says. “This nuclear body concentration will fundamentally change the dynamics of the genome organization and will very likely turn the genome from a liquid to a gel.”
This gel-like state would make it easier for different regions of the chromatin to interact with each other than if the structure existed in a liquid state, he says. Maintaining stable interactions between distant regions of the genome is important because genes are often controlled by stretches of chromatin that are physically distant from them.
The research was funded by the National Institutes of Health and the Gordon and Betty Moore Foundation.