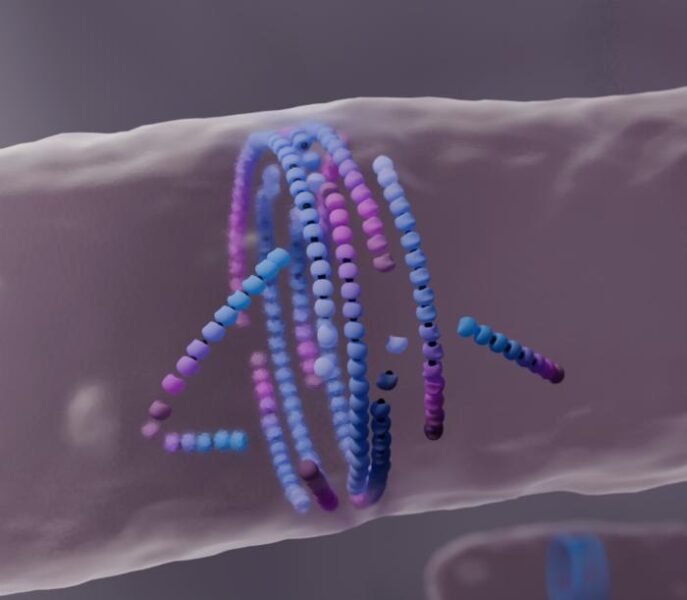
Researchers uncover how protein structures self-assemble and maintain order through a process of alignment or disassembly
A team of scientists from the Institute of Science and Technology Austria (ISTA) has discovered a novel mechanism driving bacterial cell division. The study, published in Nature Physics, sheds light on how active matter self-organizes to form essential structures within living cells.
The FtsZ Protein: A Key Player in Bacterial Cell Division
At the heart of this research is a protein called FtsZ, which forms filaments that assemble into a ring structure during bacterial cell division. This ring, known as the bacterial division ring, plays a crucial role in creating the new cell wall that separates daughter cells.
Professor Anđela Šarić, who led the study, explains the significance of their findings: “Everything in our cells is in a constant turnover. Thus, we need to start thinking of biological active matter from the prism of molecular turnover and in a way that adapts to the outside environment.”
The team developed a computational model to simulate how FtsZ proteins interact and form filaments through a process called treadmilling. This constant addition and removal of protein subunits at opposite ends of the filament allows for dynamic growth and shrinkage.
A New Paradigm: ‘Dying to Align’
What sets this research apart is the discovery of an unexpected self-assembly mechanism. The computational model revealed that when FtsZ filaments grow and collide with obstacles or become misaligned, they don’t push through like previously thought. Instead, they dissolve and “die.”
Christian Vanhille Campos, the study’s first author, describes this phenomenon: “Active matter made up of mortal filaments does not take misalignment lightly. When a filament grows and collides with obstacles, it dissolves and dies.”
This “dying to align” process leads to a local healing of the active material. As misaligned filaments disassemble, they contribute to a better overall assembly, ultimately favoring the formation of the well-aligned bacterial division ring.
The team’s findings were further validated through collaborations with experimental groups led by Séamus Holden at The University of Warwick and Martin Loose at ISTA. These partnerships allowed the researchers to confirm their computational results with real-world observations in both live bacteria and controlled laboratory settings.
Why it matters: Understanding the mechanisms behind bacterial cell division is crucial for developing new antibiotics and combating antibiotic resistance. Moreover, this research could pave the way for creating synthetic self-healing materials with applications in various fields, from medicine to materials science.
The study raises several intriguing questions for future research:
1. How does this mechanism compare to cell division processes in more complex organisms?
2. Could disrupting this alignment process be a potential target for new antimicrobial therapies?
3. How might this self-organizing principle be applied to create artificial cells or smart materials?
As the field of active matter research continues to evolve, studies like this one provide valuable insights into the fundamental processes that govern life at the cellular level. The next phase of this research, supported by a 3.7 million Euro grant from the Wellcome Trust, will focus on modeling how the bacterial division ring assists in building the cell wall that divides the cell.
By uncovering the intricate dance of protein filaments during cell division, scientists are not only advancing our understanding of basic biology but also opening doors to innovative applications in synthetic biology and materials science. The principle of “align or die” may prove to be a powerful concept in designing future self-assembling and self-healing systems.