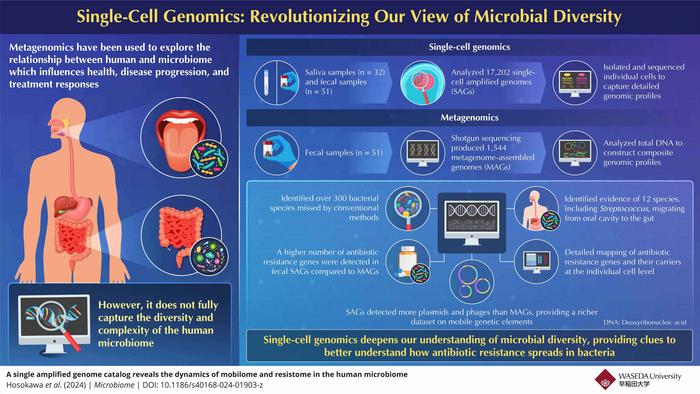
A groundbreaking study led by Associate Professor Masahito Hosokawa from Waseda University has revolutionized our understanding of the human microbiome. Using advanced single-cell genomics, researchers have uncovered hundreds of previously unknown bacterial species and mapped antibiotic resistance in unprecedented detail.
Summary: Scientists have developed a novel single-cell genome technique that reveals new bacterial species and tracks antibiotic resistance genes, offering potential breakthroughs in personalized medicine and public health strategies.
Estimated reading time: 6 minutes
The human body is home to trillions of microorganisms, collectively known as the microbiome. This complex ecosystem plays a crucial role in our health, influencing everything from digestion to immune response. However, traditional methods of studying these microbes have left many questions unanswered.
Enter single-cell genomics. This cutting-edge approach, developed by a team led by Associate Professor Masahito Hosokawa from Waseda University in collaboration with bitBiome, Inc., allows scientists to examine individual bacterial cells with unprecedented precision.
“The limitation of metagenomics inspired us to develop a new approach to explore the human microbiome at the single-cell level,” says Hosokawa. “This single-cell genome approach can enhance our understanding of how bacteria interact and exchange genetic material including antibiotic resistance genes, providing deeper insights into human health and disease.”
Unlocking the Microbial Black Box
The study, published in Microbiome on October 2, 2024, analyzed saliva and fecal samples from 51 participants using a novel technique called SAG-gel technology, commercialized as bit-MAP® by bitBiome, Inc. This method encapsulates individual bacteria in gel beads, allowing for the amplification and analysis of their genomes one by one.
The results were staggering. Researchers recovered genomes of 300 bacterial species that had been missed by conventional methods. But the discoveries didn’t stop there.
“Our study analyzed 30,000 individual genomes of oral and intestinal bacteria, which is the world’s largest genome dataset, showcasing the power of single-cell genomics in elucidating microbial diversity and interactions,” Hosokawa explains.
This massive dataset provided unprecedented insights into:
- Antibiotic resistance genes
- Gene exchange networks
- Bacterial interactions
- Microbial diversity
Battling the Superbug Threat
One of the most significant findings of the study was the detailed mapping of antibiotic resistance genes. This information could be crucial in the fight against “superbugs” – bacteria that have developed resistance to multiple antibiotics.
The single-cell approach allowed researchers to track how these resistance genes move between different bacterial species, potentially identifying key players in the spread of antibiotic resistance. This knowledge could lead to more targeted and effective treatment strategies, ultimately improving public health outcomes.
Beyond the Gut: Wide-Ranging Applications
The implications of this research extend far beyond the human body. Hosokawa notes, “Our approach provides clues to better understand how antibiotic resistance spreads in bacteria and has potential for future medical and public health applications.”
Potential applications include:
- Environmental monitoring: Tracking genetic shifts across ecosystems to manage and prevent the spread of antibiotic resistance.
- Agriculture: Guiding practices to minimize resistance spread through soil, water, and livestock.
- Personalized medicine: Tailoring treatments based on an individual’s unique microbiome profile.
Challenges and Future Directions
While the results are promising, the technology is still in its early stages. Scaling up the process to analyze larger populations and integrating the findings into clinical practice will require further research and development.
Additionally, as with any powerful new technology, ethical considerations surrounding privacy and data use will need to be addressed.
Despite these challenges, the potential benefits are immense. As we continue to unravel the complexities of the human microbiome, single-cell genomics may prove to be a key tool in our arsenal against disease and in our quest for better health.
Quiz: Test Your Knowledge
- What new technique did researchers use to study individual bacterial cells?
- How many previously unknown bacterial species were discovered using this method?
- What potential application of this research could help prevent the spread of antibiotic resistance in agriculture?
Answers:
- SAG-gel technology (commercialized as bit-MAP®)
- 300
- Guiding practices to minimize resistance spread through soil, water, and livestock
Further Reading
Glossary of Terms
- Microbiome: The collection of all microorganisms living in association with the human body.
- Single-cell genomics: A method of analyzing the genetic material from individual cells.
- Antibiotic resistance: The ability of bacteria to survive exposure to antibiotics designed to kill them.
- Metagenomics: The study of genetic material recovered directly from environmental samples.
- SAG-gel technology: A technique that encapsulates individual cells in gel beads for genomic analysis.
- Mobilome: The complete set of mobile genetic elements in a genome.
Enjoy this story? Get our newsletter! https://scienceblog.substack.com/