he slightest variation in a sequence of DNA can have profound effects. Modern genomics has shown that just one mutation can be the difference between successfully treating a disease and having it spread rampantly throughout the body.
Now, researchers have developed a new method that can look at a specific segment of DNA and pinpoint a single mutation, which could help diagnose and treat diseases such as cancer and tuberculosis. These small changes can be the root of a disease or the reason some infectious diseases resist certain antibiotics. The findings were published online this week (July 28) in the journal Nature Chemistry.
“We’ve really improved on previous approaches because our solution doesn’t require any complicated reactions or added enzymes, it just uses DNA,” said lead author Georg Seelig, a University of Washington assistant professor of electrical engineering and of computer science and engineering. “This means that the method is robust to changes in temperature and other environmental variables, making it well-suited for diagnostic applications in low-resource settings.”
DNA is a type of nucleic acid, the biological molecule that gives all living things their unique genetic signatures. In a double strand of DNA, known as a double helix, a series of base pairs bond and encode our genetic information. As genomics research has progressed, it’s clear that a change of just one base pair – a sequence mutation, an insertion or a deletion – is enough to trigger major biological consequences. This could explain the onset of disease, or the reason some diseases don’t respond to usual antibiotic treatment.
Take, for example, tuberculosis – a disease that’s known to have drug-resistant strains. Its resistance to antibiotics often is due to a small number of mutations in a specific gene. If a person with tuberculosis isn’t responding to treatment, it’s likely because there is a mutation, Seelig said.
Now, researchers have the ability to check for that mutation preventatively.
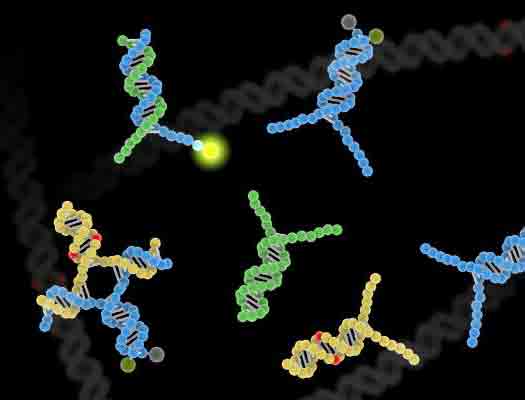
Seelig, along with David Zhang of Rice University and Sherry Chen, a UW doctoral student in electrical engineering, designed probes that can pick out mutations in a single base pair in a target stretch of DNA. The probes allow researchers to look in much more detail for variations in long sequences – up to 200 base pairs – while current methods can detect mutations in stretches of up to only 20.
“In terms of specificity, our research suggests that we can do quadratically better, meaning that whatever the best level of specificity, our best will be that number squared,” said Zhang, an assistant professor of bioengineering at Rice University.
The testing probes are designed to bind with a sequence of DNA that is suspected of having a mutation. The researchers do this by creating a complimentary sequence of DNA to the double-helix strand in question. Then, they allow molecules containing both sequences to mix in a test tube in salt water, where they naturally will match up to one another if the base pairs are intact. Unlike previous technologies, the probe molecule checks both strands of the target double helix for mutations rather than just one, which explains the increased specificity.
The probe is engineered to emit a fluorescent glow if there’s a perfect match between it and the target. If it doesn’t illuminate, that means the strands didn’t match and there was in fact a mutation in the target strand of DNA.
The researchers have filed a patent on the technology and are working with the UW Center for Commercialization. They hope to integrate it into a paper-based diagnostic test for diseases that could be used in parts of the world with few medical resources.