While human families are easily illustrated as a tree, bacterial families look more like a heap of branches. Scientists are trying to trace the connections between those branches in an effort to learn more about the bacteria that harm us, and those that do not.
UConn’s Peter Gogarten and Joerg Graf recently set out with a team of researchers to sketch family trees of 56 strains of bacteria in the Aeromonas genus, a diverse group of bacteria that live primarily in water and in the guts of blood-feeding animals such as leeches, mosquitoes, and vampire bats, but also cause disease in humans, fish, and other animals.
Through an examination of the relationships between species of bacteria in the Aeromonas genus, the researchers hoped to find clues as to which species are harmless and which are pathogenic.
Using a cutting-edge technique that compared large swaths of the Aeromonas microbial genetic code, family relationships between the bacterial strains began to emerge – but the branches of Aeromonas’ family tree are twisted indeed.
Breaking Down the Problem
Trying to piece together any bacterial family tree is complicated by the way they reproduce. Bacteria clone themselves, with each daughter bacteria an identical copy of the mother, which would seem to make tracing family lines easy. But it’s complicated by the way bacteria have sex. Sex – or the exchange and recombination of genetic material – isn’t related to the cloning process of bacterial reproduction. Instead, bacteria swap genes back and forth promiscuously all the time, even with bacteria that aren’t members of their own species.
Bacterial gene swapping, called horizontal gene transfer by microbiologists, is like a messy game of telephone, during which the information can get garbled as it passes from one bacterium to another. Gene swapping makes it particularly difficult to trace species evolution. Microbiologists routinely argue over whether two strains are in the same species or not, and some will tell you straight that the whole concept of bacterial species is suspect.
“People still struggle with the species concept in bacteria,” says microbiologist Joerg Graf. “Traditionally, 70 percent identical DNA shared between two bacterial organisms means they are in the same species.”
Graf is a believer in bacterial species, while his colleague Peter Gogarten has doubts. But the two agree that the question is worth investigating, because you need to know what you’re dealing with, and species is as good a label as any.
The UConn researchers, along with collaborators at the University of Montpelier in France, show in the Nov. 18 issue of mBio [[ http://mbio.asm.org/content/5/6/e02136-14.full ]] that they can effectively group different strains of bacteria into species using genomic analysis.
The researchers compared the genomes of the 56 strains using two computerized techniques. One bioinformatic technique they used mimicked an older, messy lab procedure called DNA-DNA hybridization that was the gold standard for this kind of work for many years. The other one was Average Nucleotide Identity (ANI), a cutting-edge side-by-side comparison of large chunks of DNA.
The results clearly grouped many strains of Aeromonas into distinct species, and showed that several strains of Aeromonas bacteria in the GenBank, a collection of publicly available genetic sequences held by the National Institutes of Health, are misnamed. In fact, two of them appear to be different enough from other Aeromonas to qualify as new species entirely.
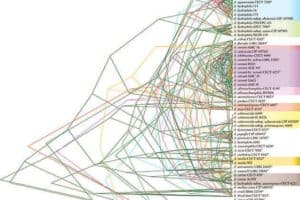
The researchers also traced 16 “housekeeping genes,” important for survival, through each of the 56 strains. They tracked how the genes shifted position through the genome and changed, and used those changes to track when each lineage of bacteria split off from one another. For each gene, the researchers were able to create a phylogenetic or ‘family’ tree that grouped the Aeromonas into species, and then showed how those species were related to each other. Species that shared identical or very close versions of a housekeeping gene could be thought of as siblings, while species with quite different versions of that gene were more like distant cousins. Except there was a problem.
“None of those trees agreed,” says Gogarten.
Stumped
The phylogenetic trees weren’t wrong, exactly; they all agreed quite closely on which strains of bacteria were in the same species. It was in the deeper relationships, the way the bacteria were related to other species in the Aeromonas genus, where the trees began to differ. The bacteria’s constant game of genetic telephone seems to have included even these essential genes.
The researchers had been hoping to see a pattern. Perhaps one branch had all the disease-producing agents for humans and another one had all the fish pathogens, for example. The researchers could then look for genes that help Aeromonas sicken humans or fish, or perhaps predict whether a newly discovered species could cause disease. But no such luck. The gene trees were all mixed up between pathogenic and harmless bacteria.
Gogarten and Graf say they may never be able to untangle Aeromonas’ family tree, but this work shows that their bioinformatic techniques are good. Their next step will be to analyze the Aeromonas genomes to find the genes that allow them to survive in specialized environments like the gut of a vampire bat or human blood vessels.
They have also set up a website, Aeromonasgenomes.uconn.edu, where other researchers can access known genomes for Aeromonas and post ones they’ve sequenced themselves.