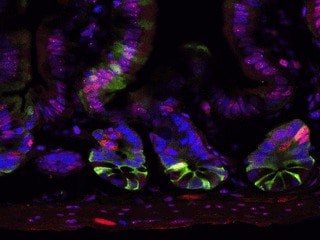
The suckling period (infancy) in mice is critical for epigenetic changes (changes that affect the way genes are expressed) in the development of stem cells in the intestine, potentially affecting intestinal health for life. Moreover, the intestinal microbiome guides these epigenetic processes, said researchers at the USDA/ARS Children’s Nutrition Research Center at Baylor College of Medicine and Texas Children’s Hospital in a report that appears today in the journal Genome Biology.
“In humans, the gastrointestinal tract (or gut) can be affected by many diseases, including inflammatory bowel disease and colorectal cancer,” said Dr. Lanlan Shen, associate professor of pediatrics at Baylor and senior author of the study.
“Cells are continually being replaced in the intestinal epithelium. They only last around four days and then new cells are formed,” said Dr. Robert Waterland, associate professor of pediatrics at Baylor, who was also involved in the study. “It’s a very dynamic process. The cells that control this are called intestinal epithelial stem cells and these are the cells that have the ability to continually make new intestinal epithelium cells that our body uses. These stem cells are essentially the control center of the gut. These are the cells that are going to regulate your gut physiology for your entire life.”
“Using mouse models, we were able to isolate and study a pure stem cell population,” said Shen. When the researchers used whole genome sequencing to study these cells during the suckling and weaning periods, they found that DNA methylation (the addition of a methyl molecule) plays a regulatory role in intestinal stem cells. In fact, the methylation of CpG islands (CGIs) turns on important genes involved in the development of these cells. In many cases, this epigenetic development depends on some of the bacteria that make up the gut microbiome.
While many recent studies demonstrate that the gut microbiome has a significant and long-term impact on gastrointestinal health, this new work provides a clue about how this works. In addition, researchers can use markers identified in this study and see if they can translate this to humans, perhaps even develop an epigenetic maturation index in infants to guide optimal gut health. Finally, it opens the possibility to identify early environmental causes of gastrointestinal disease, and potentially reverse them using probiotic therapy.
“This promises some exciting opportunities to understand how we might be able to tailor one’s microbiome exposure during infancy to maximize health and reduce gastrointestinal disease throughout life,” said Waterland.
Others who took part in the study include Dai-Hai Yu, Manasi Gadkari, Quan Zhou, Shiyan Yu, Nan Gao, Yongtao Guan, Deborah Schady, Tony N. Roshan, Miao-Hsueh Chen, Eleonora Laritsky, Zhongqi Ge, Hui Wang, Rui Chen, , Chelsea Moriarty, Cindy Hwang and Alton G. Swennes of Baylor; Caroline Westwater of the Medical University of South Carolina, Lynn Bry of Brigham and Women’s Hospital Harvard Digestive Diseases Center; and Sean R. Moore of Cincinnati Children’s Hospital Medical Center.
This work was supported by grants from the Sidney Kimmel Foundation, US Department of Agriculture, March of Dimes, the National Institutes of Health and a Bill & Melinda Gates Grand Challenges Explorations Grant.