Biochemists have long known that crucial cell processes depend on a highly regulated cleanup system known as proteolysis, where specialized proteins called proteases degrade damaged or no-longer-needed proteins. These proteases must destroy their specific targets without damaging other proteins, but how this orderly destruction works is unknown in many cases.
Now researchers in Peter Chien’s group in the department of biochemistry and molecular biology at the University of Massachusetts Amherst report finding how an essential bacterial protease controls cell growth and division. Details appear in the current issue of CELL.
Lead author Kamal Joshi, a doctoral candidate in the Chien lab, conducted experiments in the model bacterium Caulobacter crescentus. In this species, the ability to grow and replicate DNA is regulated by ClpXP, a highly conserved protease that in many bacteria allows them to cope with stressful environments such as the human body. Understanding how ClpXP is controlled could open a path to antibiotics that inhibit harmful bacteria in new ways.
Chien explains that one of the long-standing mysteries in the field is how proteolysis controls Caulobacter growth. “The odd thing is that ClpXP is always present during different growth stages, but only destroys its target proteins at a specific time,” he says. This led his group to think, “There must be a missing factor that we couldn’t see, because something has to come on the scene to accomplish this. There was some genetic evidence pointing to certain additional proteins, but we didn’t know how they worked.”
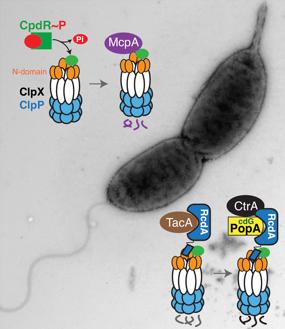
His lab took a biochemical approach to the problem, purifying all available proteins and designing experiments to query how they interacted and what functions were affected in their presence or absence. In this way, “Kamal found that the ClpXP protease could not by itself destroy the target proteins,” Chien says. Instead, “he found that the additional regulatory proteins we had detected were controlling different parts of the process.”
Further, Joshi found that these newly identified regulatory adaptors worked in a step-wise hierarchical way. The first adaptor was directly responsible for degrading a handful of proteins, but it could also recruit an additional adaptor that would deliver a different set of proteins and bind even more adaptors. Working with the Viollier lab from the University of Geneva, Switzerland, the researchers found scores of additional protease targets that were destroyed in this hierarchical way.
The researchers believe this new fundamental knowledge may offer an entirely new target for developing new antibiotics with a high potential to avoid triggering drug resistance. This is because new compounds could be devised which would not simply target all bacterial growth, but only a specific pathway, such as virulence.
Such a protease-based technique would be very specific and less likely to generate the global selective pressure that leads bacteria to develop resistance. Chien points out, “In this approach, you don’t force the benign bacteria to develop resistance because their growth isn’t threatened. The hope is to target only those pathogenic organisms that are trying to overcome the stressful environment inside the human body.”
At present, the researchers do not know if these adaptor factors are common among all bacteria, but they intend to find out. “How proteases selectively destroy their targets is a universal problem,” says Chien. “We think that these same mechanisms also may be found in other bacteria, but won’t know until we start testing them.” Work in the Chien lab is funded by the NIH National Institute of General Medical Sciences.