As you read this text, the millions of cells that make up your body are hard at work. Within every cell is a flurry of activity keeping you alive, mostly driven by machinery that is made up of proteins. Some of this protein machinery is so important to living things that it has remained unchanged over millions of years of evolution.
One of these ancient cellular machines is the endoplasmic reticulum membrane protein complex (EMC), which is present in all eukaryotes, from mushrooms to insects to mice to humans. Shaped like a kind of knobby tube, the EMC acts as a gateway between the interior of the cell and the cell’s outer membrane, and behaves as a kind of border control through which only certain proteins can pass. The EMC interacts with proteins involved in a wide range of important processes, such as regulating blood pressure and heart rate, and the levels of neurotransmitters like serotonin and dopamine.
Now, in a new paper published in the July issue of the journal Molecular Cell, researchers describe how the cell assembles the EMC out of nine other smaller protein structures, or subunits, that fit together perfectly. The research was conducted in the Caltech laboratory of Rebecca Voorhees, assistant professor of biology and biological engineering and Heritage Medical Research Institute Investigator.
The cell makes each of the nine EMC subunits separately; to form the EMC, these separate proteins must find one another in the relative vastness of the cell and assemble themselves. As an analogy, imagine putting together an IKEA dresser from its separate components: drawers (that also must first be assembled), body, screws, knobs, and so on. Each piece has a precise place in the whole structure. In research published last year, the Voorhees lab created the first-ever images of the EMC with atomic-level detail, revealing the blueprint of the structure.
EMC assembly needs to happen quickly, because if a cell detects that an orphaned subunit has been hanging around too long, quality control processes will likely destroy it, in the same way you might throw out any extra screws left over from your IKEA assembly.
“The EMC is essential in all eukaryotic organisms,” says Voorhees. “If the EMC can’t assemble, then the organism will not survive.”
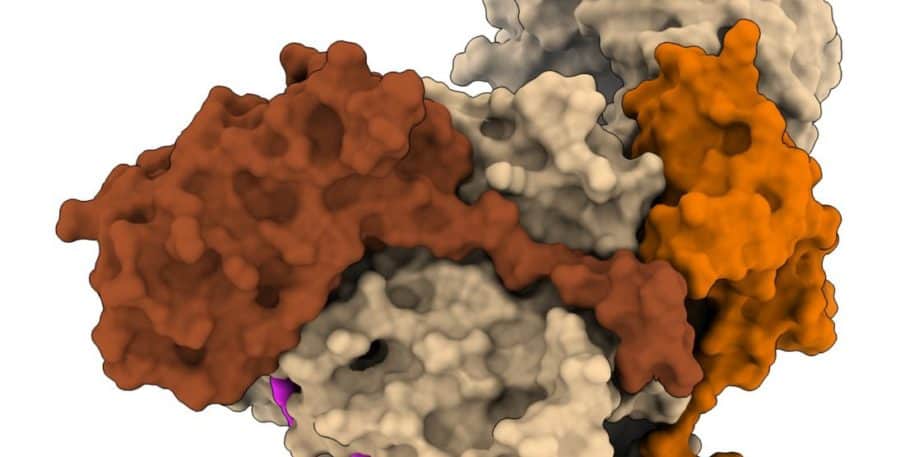
To help them come together to form the EMC, each subunit has specific locations on its surface, called binding sites, where it attaches to specific other subunit proteins. But when floating around the cell waiting to link up with its counterparts, a subunit is vulnerable to accidentally binding with other proteins also loose within the cell. As an analogy, imagine using a magnet to search for a needle in a haystack; if there are other metal objects within the haystack that can stick to your magnet and cover up its surface, preventing it from binding with the needle you seek, you might never find it. Therefore, the EMC subunits (and other proteins) need a way to shield their binding sites until they are ready to bind with the correct partners.
This is where proteins called assembly factors come in. An assembly factor protein physically covers and protects the sensitive binding sites but is not stuck there forever. It will peel off, like tape, when the subunit finds a correct protein to bind with, and then the two proteins “super-glue” together at the binding site.
This multistep assembly process is a major research focus for the Voorhees laboratory. In the new study, the team aimed to determine how the EMC correctly assembles. The group focused on one subunit in particular, EMC2, which serves as the backbone of the EMC complex architecture, as it binds to six of the other eight EMC subunits.
In this work, the team found that another protein, called WNK1, acts as the assembly factor for EMC2, temporarily shielding its sensitive binding sites until it could rendezvous with other EMC subunits. This was surprising: Although WNK1 is known to play several important roles in the cell (mutations in WNK1 lead to congenital hypertension, for example), there had been no indication that it would protect the binding sites on EMC2.
WNK1 is a relatively large protein, and most researchers have focused on studying a small region on the protein called a kinase, which catalyzes reactions. But WNK1 also has a long “tail,” the purpose of which was not understood. This new research shows that part of the tail is responsible for covering EMC2’s binding site.
“People have studied the kinase domain for years now, but no one really knew what WNK1’s tail was doing,” Voorhees explains. “The tail is about 10 times heavier than the kinase region, so it’s huge in comparison to the part we understand. It was surprising for us to discover that one piece of the tail is what is binding EMC2. It was a piece that had never been previously ascribed to have any function, but it is found throughout all higher eukaryotes. In future work, we’re really interested to understand why WNK1 is involved in the EMC in this way.”
The paper is titled, “WNK1 is an assembly factor for the human ER membrane protein complex.” The first author is postdoctoral fellow Tino Pleiner. In addition to Pleiner and Voorhees, co-authors are graduate students Masami Hazu and Giovani Pinton Tomaleri, former staff scientist Kurt Januszyk, lab manager Robert Oania, former senior bioinformatician Michael Sweredoski, former senior lab manager Annie Moradian, and former visiting researcher Alina Guna. Funding was provided by the Heritage Medical Research Foundation, the Kinship Foundation, the Pew-Stewart Foundation, and the National Institute of General Medical Sciences of the National Institutes of Health.