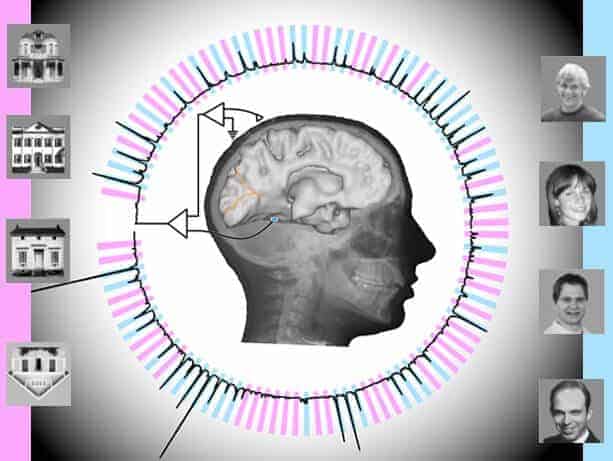
Using electrodes implanted in the temporal lobes of awake patients, scientists have decoded brain signals at nearly the speed of perception. Further, analysis of patients’ neural responses to two categories of visual stimuli – images of faces and houses – enabled the scientists to subsequently predict which images the patients were viewing, and when, with better than 95 percent accuracy.
The research is published today in PLOS Computational Biology.
University of Washington computational neuroscientist Rajesh Rao and UW Medicine neurosurgeon Jeff Ojemann, working their student Kai Miller and with colleagues in Southern California and New York, conducted the study.
“We were trying to understand, first, how the human brain perceives objects in the temporal lobe, and second, how one could use a computer to extract and predict what someone is seeing in real time?” explained Rao. He is a UW professor of computer science and engineering, and he directs the National Science Foundation’s Center for Sensorimotor Engineering, headquartered at UW.
“Clinically, you could think of our result as a proof of concept toward building a communication mechanism for patients who are paralyzed or have had a stroke and are completely locked-in,” he said.
The study involved seven epilepsy patients receiving care at Harborview Medical Center in Seattle. Each was experiencing epileptic seizures not relieved by medication, Ojemann said, so each had undergone surgery in which their brains’ temporal lobes were implanted – temporarily, for about a week – with electrodes to try to locate the seizures’ focal points.
“They were going to get the electrodes no matter what; we were just giving them additional tasks to do during their hospital stay while they are otherwise just waiting around,” Ojemann said.
Temporal lobes process sensory input and are a common site of epileptic seizures. Situated behind mammals’ eyes and ears, the lobes are also involved in Alzheimer’s and dementias and appear somewhat more vulnerable than other brain structures to head traumas, he said.
In the experiment, the electrodes from multiple temporal-lobe locations were connected to powerful computational software that extracted two characteristic properties of the brain signal: “event-related potentials” and “broadband spectral changes.”
Rao characterized the former as likely arising from “hundreds of thousands of neurons being co-activated when an image is first presented,” and the latter as “continued processing after the initial wave of information.”
The subjects, watching a computer monitor, were shown a random sequence of pictures – brief (400 millisecond) flashes of images of human faces and houses, interspersed with blank gray screens. Their task was to watch for an image of an upside-down house.
“We got different responses from different (electrode) locations; some were sensitive to faces and some were sensitive to houses,” Rao said.
The computational software sampled and digitized the brain signals 1,000 times per second to extract their characteristics. The software also analyzed the data to determine which combination of electrode locations and signal types correlated best with what each subject actually saw.
In that way it yielded highly predictive information.
By training an algorithm on the subjects’ responses to the (known) first two-thirds of the images, the researchers could examine the brain signals representing the final third of the images, whose labels were unknown to them, and predict with 96 percent accuracy whether and when (within 20 milliseconds) the subjects were seeing a house, a face or a gray screen.
This accuracy was attained only when event-related potentials and broadband changes were combined for prediction, which suggests they carry complementary information.
“Traditionally scientists have looked at single neurons,” Rao said. “Our study gives a more global picture, at the level of very large networks of neurons, of how a person who is awake and paying attention perceives a complex visual object.”
The scientists’ technique, he said, is a steppingstone for brain mapping, in that it could be used to identify in real time which locations of the brain are sensitive to particular types of information.
Lead author of the study is Kai Miller, a neurosurgery resident and physicist at Stanford University who obtained his M.D. and Ph.D. at the UW. Other collaborators were Dora Hermes, a Stanford postdoctoral fellow in neuroscience, and Gerwin Schalk, a neuroscientist at the Wadsworth Institute in New York.
“The computational tools that we developed can be applied to studies of motor function, studies of epilepsy, studies of memory. The math behind it, as applied to the biological, is fundamental to learning,” Ojemann said.
This work was supported by National Aeronautics and Space Administration Graduate Student Research Program, the National Institutes of Health (R01-NS065186, T32-EY20485, R01-EB00856 and P41-EB018783), the National Science Foundation (EEC-1028725), and the U.S. Army.
If our reporting has informed or inspired you, please consider making a donation. Every contribution, no matter the size, empowers us to continue delivering accurate, engaging, and trustworthy science and medical news. Independent journalism requires time, effort, and resources—your support ensures we can keep uncovering the stories that matter most to you.
Join us in making knowledge accessible and impactful. Thank you for standing with us!