Human cells make new copies of their DNA billions of times each day, a crucial process upon which life itself depends. However, scientists do not fully understand how cells unzip the double-stranded DNA molecule before replicating both halves of it. New work at Rockefeller may help change that.
For the first time, researchers in Michael O’Donnell’s Laboratory of DNA Replication have built a model that can enable scientists to study what happens at the “replication fork” — the point where the DNA molecule is split down the middle in order to create an exact copy of each side.
“As a research tool, our model could help scientists better address basic biological questions about cell division, as well as the nature of errors that cause diseases, such as cancer,” O’Donnell says. “There are plenty of hypotheses about the mechanics of DNA replication, but until now the process could not be studied using a defined system with pure proteins.”
The findings, published July 6 in the journal Nature Structural and Molecular Biology, focus on eukaryotic cells. All multicellular life, including humans, is eukaryotic. Single-celled prokaryotes evolved a different method for replicating their DNA. In order to study the replication fork, O’Donnell and his laboratory needed to recreate the process in a simple model. In a test tube, they brought together a set of DNA building blocks known as nucleotides, a double-stranded molecule of DNA and the enzymes essential to the process. These enzymes came from the yeast fungus, which is a eukaryote.
According to O’Donnell, the team’s technique may allow researchers to reconstruct biochemical events that were, until now, difficult or impossible to study in detail. For example, scientists know some inheritable information, known as epigenetic information, is not encoded in the DNA, but instead lies in modifications to proteins associated with the DNA. Yet exactly how this information is passed on remains a mystery. Scientists also don’t know what happens when the replication fork encounters an area of damaged DNA as it travels down the length of the molecule.
These processes have direct implications for the study of diseases, such as cancer, that can arise from DNA damage or problems with epigenetic inheritance.
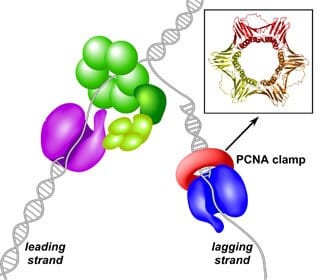
Scientists already know the replication fork is assembled as a complex of numerous proteins that unwinds and separates the DNA into two individual strands. The emerging replication fork looks much like a zipper opening, with a protein complex in the role of a zipper slider and the two strands of the separating DNA molecule appearing like the two rows of teeth of the open zipper. Each side of the zipper becomes the template for a daughter strand, but the mechanics differ depending on the side.
The researchers in O’Donnell’s lab took a close look at this essential asymmetry, which arises because the two strands of double-stranded DNA fit together head-to-tail. But since replication can only progress in one direction, the two daughter strands — one lagging, one leading — are put together at slightly different paces and in opposite directions. Each strand relies on a different enzyme, and, using the new yeast-based model, the researchers were able to explore how these two very different enzymes attach to the DNA in order to replicate it.
“I believe this new tool opens up replication-fork biology to biochemical study by our own and many other labs, providing a new tool to unravel some pressing questions in a number of fields of study, including epigenetics and DNA repair,” O’Donnell says.
If our reporting has informed or inspired you, please consider making a donation. Every contribution, no matter the size, empowers us to continue delivering accurate, engaging, and trustworthy science and medical news. Independent journalism requires time, effort, and resources—your support ensures we can keep uncovering the stories that matter most to you.
Join us in making knowledge accessible and impactful. Thank you for standing with us!