Plant biologists have sequenced the genome of a particularly promising species of green alga, providing a blueprint for new discoveries in producing sustainable biofuels, antioxidants, and other valuable bioproducts.
The researchers targeted Chromochloris zofingiensis, a single-celled green alga that has drawn commercial interest as one of the highest producers of the best lipids for biofuel production.
The team of scientists, led by researchers at the Department of Energy’s (DOE) Lawrence Berkeley National Laboratory (Berkeley Lab) in collaboration with the University of California, Los Angeles, recently published their work in the Proceedings of the National Academy of Sciences.
The work was conceived of and developed at Berkeley Lab by Krishna Niyogi, faculty scientist and an investigator at the Howard Hughes Medical Institute.
“This genome will be an important resource to develop renewable and sustainable microalgal biofuels to facilitate clean energy and a cleaner environment,” said study lead author Melissa Roth, a postdoctoral researcher in Niyogi’s lab. “Algae absorb carbon dioxide and are intrinsically solar-powered by photosynthesis, but C. zofingiensis has an added benefit in that it can be cultivated on non-arable land and in wastewater.”
Niyogi also pointed out that C. zofingiensis is a natural source for astaxanthin, an antioxidant derived from dietary algae that gives salmon its pinkish hue. In algae, astaxanthin is thought to provide protection from oxidative stress.
“This alga has potential as a nutraceutical,” said Niyogi, who is also a UC Berkeley professor of plant and microbial biology. “Studies are already underway to determine whether astaxanthin’s anti-inflammatory properties are beneficial in treatments for cancer, cardiovascular disease, neurodegenerative disease, diabetes, and other human health problems.”
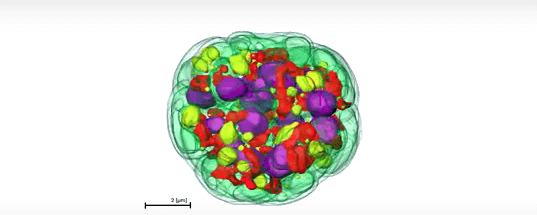
To get an inside look at the cells, the researchers relied upon the National Center for X-ray Tomography (NCXT), a joint Berkeley Lab-UCSF program located at the Lab’s Advanced Light Source. Using soft X-ray tomography, a technique comparable to a computerized tomography scan, scientists imaged and then reconstructed sections of the algal genome to generate a 3-D view. Cells were captured dividing into two, four, and even sixteen daughter cells.
“Combining multiple sequencing techniques, we were able to generate a chromosome-level assembly of the genome, which is an uncommonly high level of architecture for an alga and similar to that of a model organism. In fact, the quality of the C. zofingiensis genome rivals the model green alga Chlamydomonas reinhardtii, which was first sequenced about a decade ago,” said Roth.
The alga contains approximately 15,000 genes.
Other senior authors on the paper include Sabeeha Merchant, UCLA professor of biochemistry; Matteo Pellegrini, UCLA professor of molecular, cell, and developmental biology; and Carolyn Larabell, NCXT director and professor of anatomy at UCSF.
This research was supported by DOE’s Office of Science, the US Department of Agriculture, the National Institute of General Medical Sciences, and the Gordon and Betty Moore Foundation. The Advanced Light Source is a DOE Office of Science User Facility. NCXT is jointly funded by DOE and the National Institutes of Health.
If our reporting has informed or inspired you, please consider making a donation. Every contribution, no matter the size, empowers us to continue delivering accurate, engaging, and trustworthy science and medical news. Independent journalism requires time, effort, and resources—your support ensures we can keep uncovering the stories that matter most to you.
Join us in making knowledge accessible and impactful. Thank you for standing with us!